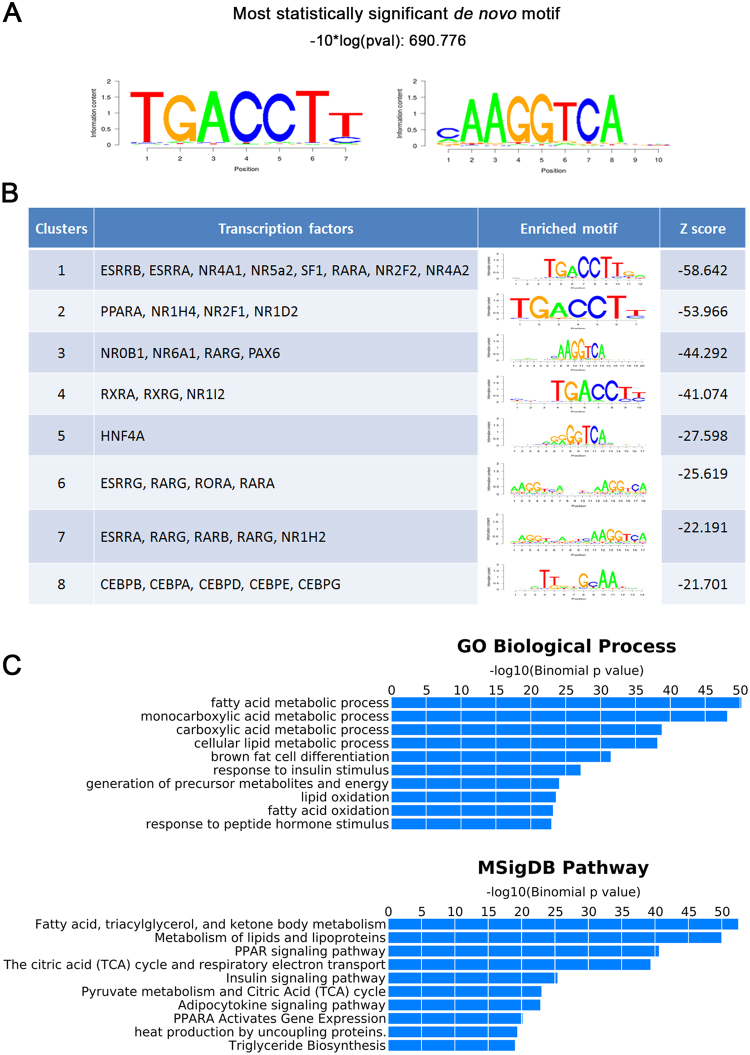
Figure 3.
Functional annotation of PGC-1α/NT-PGC-1α ChIP-seq peaks. (A) The highest scoring sequence motif compiled from PGC-1α/NT-PGC-1α binding peaks within a 600-bp window centered on the binding summits. (B) Transcription factor binding enrichment from the PGC-1α/NT-PGC-1α ChIP-seq. The top eight most enriched motifs identified using the SeqPos motif tool from Galaxy Cistrome. ESRRB (ERR beta), ESRRA (ERR alpha), NR4A1 (NUR77, TR3), NR5A2 (LRH-1, FTF), NR2F2 (TFCOUP2), NR4A2 (NURR1), NR1H4 (FXR), NR2F1 (COUP-TF1), NR1D2 (Rev-Erb), NR0B1 (DAX1), NR6A1 (RTR), NR1I2 (PXR), ESRRG (ERR gamma), and NR1H2 (LXRB). (C) Functional enrichment analysis of PGC-1α/NT-PGC-1α peaks using GREAT. Top ten ranked biological processes (GO biological process) and signaling pathways (MsigDB pathway) of genes associated with PGC-1α and NT-PGC-1α peaks are shown.