Fig. 6.
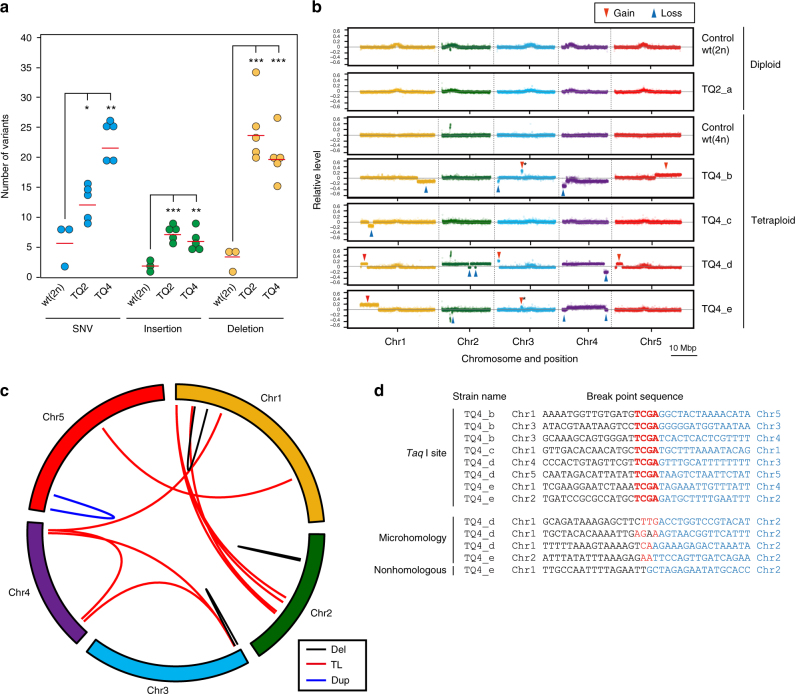
TaqI-induced large copy number variations (CNVs) and chromosome rearrangements in Arabidopsis plants. a Number of SNVs and InDels in TAQed diploid (TQ2) and tetraploid (TQ4) progenies. Data represent the number of individual variants (dots) and the mean (red line) *P < 0.05, **P < 0.01, ***P < 0.001; Welch’s t-test. b Array comparative genome hybridization (aCGH) karyotyping of TAQed diploid (TQ2_a) and tetraploid (TQ4_b-e) Arabidopsis plants with large CNVs and control plants [wt(2n) and wt(4n)]. Y axis exhibit relative fluorescence intensity of log10 (Cy5[sample]/Cy3[control]). Large CNVs (gain) in the upper arm of Chr3, indicated by an asterisk, were excluded from subsequent analysis because they occurred independently in multiple progenies. c Circular ideogram showing all TLs and large deletions identified in four TQ4 strains. d Breakpoint sequences in TAQed tetraploids with large CNV events. Consensus sequences are shown in red