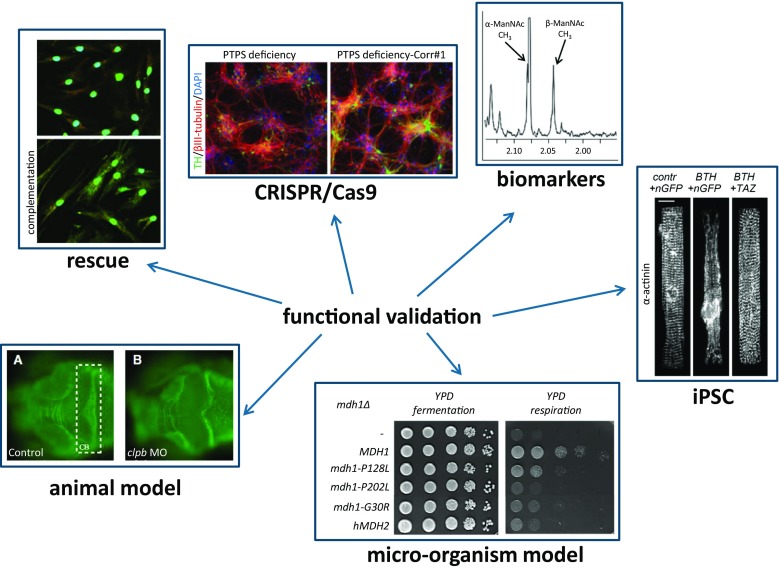
Fig. 2.
Examples of functional genomics approaches. These approaches, or combinations thereof, are frequently used to investigate the pathogenicity of genetic variants of unknown clinical significance. The examples shown are described in a clockwise order starting in the top-left corner. Rescue: introduction by lentiviral transduction of wild type LYRM7 cDNA in fibroblasts from a patient with a defect in LYRM7 results in normalization of mitochondrial Rieske Fe-S protein (Hempel et al. 2017). CRISPR/Cas9: Absence of thymidine hydroxylase (TH) staining in iPSC-derived dopaminergic nerve cells from a patient with a defect in PTPS, and normalization of TH expression after CRISPR/Cas9-mediated correction of the PTPS gene (Ishikawa et al. 2016). Biomarkers: detail of a 1H NMR spectrum of a CSF sample from a NANS patient, showing the presence of alpha and beta forms of N-acetylmannosamine (van Karnebeek et al. 2016). iPSC: Abnormal sarcomere organization in iPSC-cardiomyocytes derived from fibroblasts from a Barth-syndrome patient (BTH) with mutations in the tafazzin gene (TAZ), and normalization of sarcomeres after transfection with TAZ-mRNA (Wang et al. 2014). Micro-organism model: Restoration of growth on a non-fermentable carbon source (YPG) of a mitochondrial malate dehydrogenase-deficient yeast strain (mdh1Δ) by transfection with wild type MDH1, but not with various mdh-mutants (Ait-El-Mkadem et al. 2017). Animal model: A cerebellar defect in CLPB knock-down zebrafish embryos, as seen in Ac-tubulin stained embryos (the cerebellum is indicated by the rectangle in the control animal) (Wortmann et al. 2015, 2015)