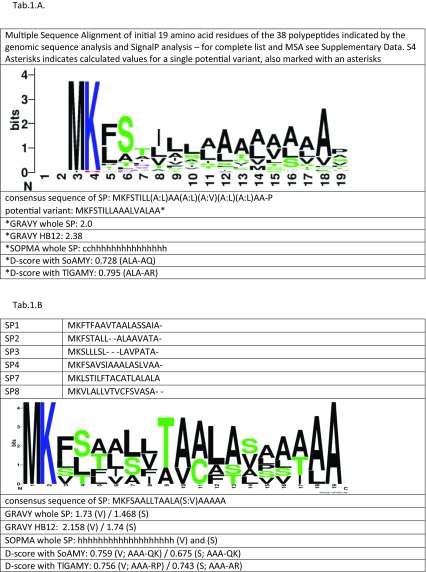
Table 1.
Signal peptide consensus sequence logos, secondary structure prediction, hydrophobicity assessment, and D score values for (a) the 38 proteins identified through Y. lipolytica CLIB122 genomic sequence scan for ORFs bearing MK x{7–15} (x,A/P){2–10} x{10–120} KR motif and (b) SPs having the highest secretory capacity in Y. lipolytica cells with SoAMY and TlGAMY polypeptides
Amino acid sequences were aligned using MEGA 7.0.14 package and ClustalW algorithm; dashes indicate gaps; ambiguous sites are indicated as (X:Y) X and Y—alternative amino acid residues occurring with the same frequency. The logos were created using Web Logo tool. Color code: black—hydrophobic amino acid residue, blue—positively charged amino acid residue, green—polar uncharged amino acid residue. Size of a letter represents frequency of a corresponding amino acid residue occurrence in a respective position. Hydrophobicity evaluation was done using GRAVY calculator tool and secondary structure prediction—using SOPMA tool. (A) Due to relatively high sequence degeneration SOPMA and GRAVY analyses were done for one possible variant; (B) Both analyses were done for the two possible variants of the consensus sequence