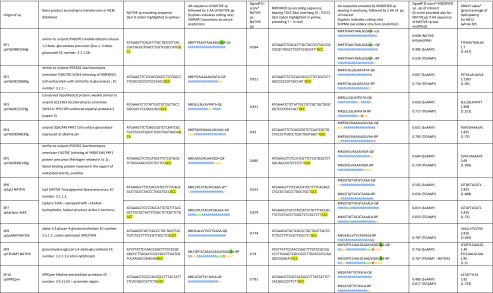
Table 2.
Signal peptides under study accompanied by the results of computational analyses
aSOPMA: prediction of secondary structure;  : alpha-helix,
: alpha-helix, : beta turn,
: beta turn,  : random coil,
: random coil,  : extended strand
: extended strand
bD score is used to discriminate signal peptides from non-signal peptides based on probability of the presence of a signal peptidase cleavage site; SignalP
cGRAVY value: grand average hydropathy calculated for 12 residues after the last positively charged residue of the n-region (HB12) or complete SP
dSignalP calculates signal peptide of 27 AA (MKLSTILFTACATLAAALPSPITPSEA-A V); PrediSi tool predicts the pre region of 17 AA (as reported in primary scientific papers (Pigniede et al., 2000), and this pre-sequence was analyzed in this study