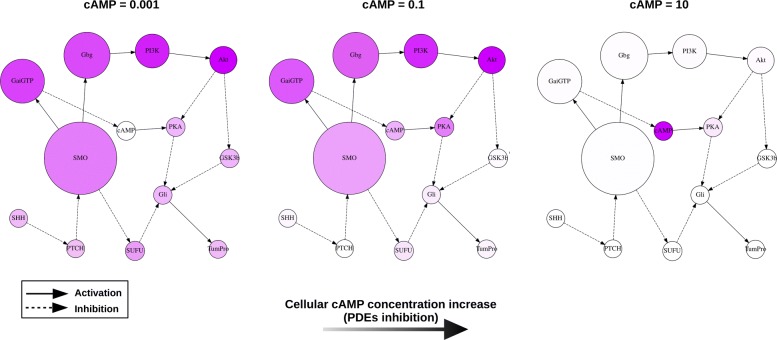
Fig. 1.
The interaction network of SMO and signalling pathway analysis. Signalling pathway analysis represents the involvement of the network components in stimulating the tumour proliferation end-point (TumPro) upon an increase in SMO independent activity (β(SMO)=0.001→0.1). See the Methods section for a description of the components. The effect of PDEs inhibition is represented as an increase in the concentration of cAMP (from left to right). The sizes and colour intensities of the nodes summarise the results of signalling pathway analysis and represent how the signal is mediated from SMO to the network end-point. The node size represents the association of a node with SMO when stimulating the end-point (the larger the node the stronger the association). The node colour indicates the signal flow (the darker the more a node can deliver a signal downstream to it)