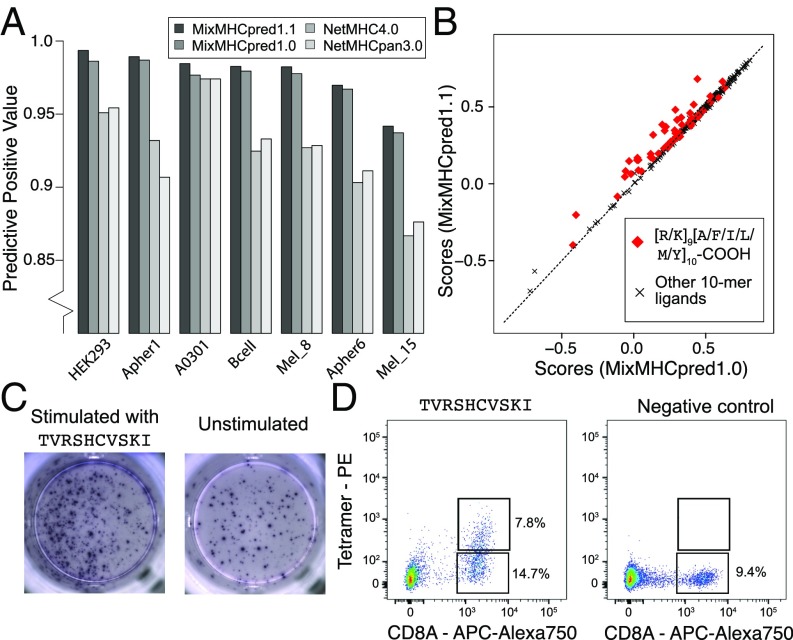
Fig. 5.
(A) Benchmarking of our HLA-I ligand predictor (MixMHCpred1.1). The y axis shows the positive predictive value among the top 20% of the predictions. (B) Analysis of scores when explicitly modeling C-terminal extensions (MixMHCpred1.1) or not (MixMHCpred1.0) for the 10-mer HLA-A03:01 ligands from a monoallelic cell line (2), as a function of the C-terminal amino acids (P9 and P10). (C) IFN-γ–ELISpot results obtained by stimulation with a C-terminally extended 10-mer HLA-A03:01 ligand (TVRSHCVSKI, from CMV, Left) vs. no peptide (Right) of a PBMC sample from a HLA-A03:01 and CMV seropositive healthy donor. (D) Multimer analysis of CD8 T cells from a healthy donor recognizing the HLA-A03:01 restricted C-terminally extended 10-mer peptide TVRSHCVSKI (Left) and the negative control (RVRAYFYSKV/HLA-A03:01 tetramer) for which we did not observe T cell recognition (Right).