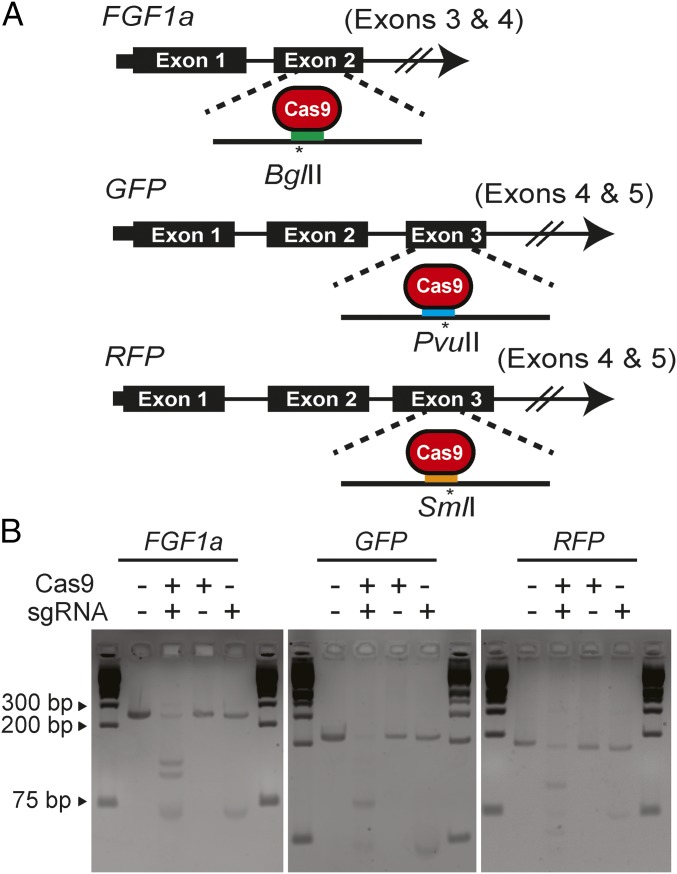
Fig. 1.
Design and activity in vitro of sgRNAs targeting A. millepora genes. (A) sgRNAs targeting exon 2 (of at least four) of FGF1a, exon 3 (of five) of GFP genes, and exon 3 (of five) of RFP genes were designed to induce double-strand breaks near endogenous restriction-enzyme sites that could be used to detect induced mutations. Colored bars, approximate locations of the sgRNA-binding sites; asterisks, predicted Cas9 cleavage sites and the nearby restriction sites. (B) Digestion in vitro of FGF1a, GFP, or RFP target DNA (SI Appendix, SI Materials and Methods) incubated with Cas9 protein, the appropriate sgRNA (as transcribed in vitro from the pDR274-based construct), or both. Fragments were analyzed by gel electrophoresis; outside lanes of each gel show molecular-size markers.