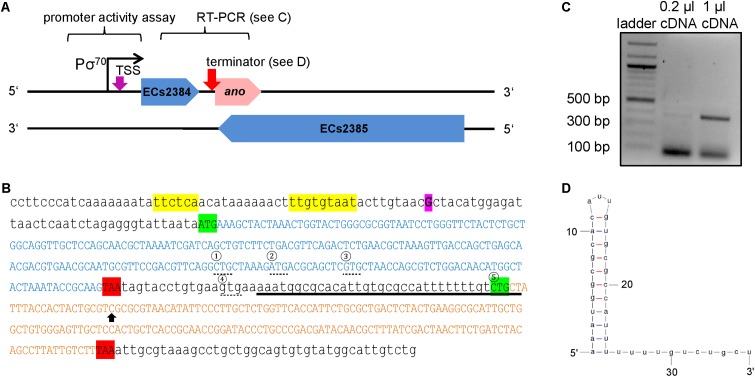
FIGURE 2.
Genomic organization of ECs2384 and ano overlapping to its mother gene ECs2385. (A) The annotated genes ECs2384 and ECs2385 are depicted in blue and the novel OLG ano is depicted in pink. The genes and the distances are drawn to scale. The predicted σ70 promoter, the experimentally determined transcription start site (TSS, purple arrow), and the predicted ρ-independent terminator between ECs2384 and ano (pink arrow) are sketched. DNA stretches, which were used for promoter activity assay and RT-PCR, are indicated. (B) DNA sequence of ECs2384 and ano. The sequence of the annotated gene ECs2384 is written in blue capital letters. The sequence of ano is written in pink capital letters. The start codons are highlighted in green and the stop codons in red. The four alternative upstream start codons of ano are marked by a dashed line and numbered consecutively. The fifth start codon, we propose as the correct one (see text). The TSS detected by 5′ RACE is highlighted in purple. The predicted σ70 promoter is colored in yellow and the predicted ρ-independent terminator is underlined. Black arrow: the nucleotide C was changed to an A in order to create a stop codon (compare to Figure 4). (C) Agarose gel picture of the RT-PCR product ECs2384-ano. A 100 bp DNA ladder (NEB) was used as a size standard. A primer pair was used for PCR with EHEC cDNA as template spanning the sequences of ECs2384 and ano. The gel shows a product of the anticipated size of 380 bp, indicating that ECs2384 and ano are transcribed as a bicistronic mRNA. (D) Secondary structure of the ρ-independent terminator predicted using Mfold.