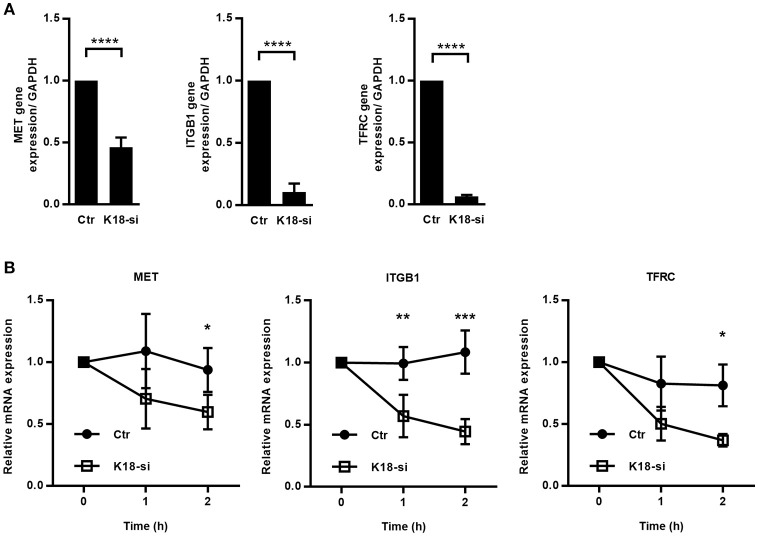
Figure 8.
K18 favors expression of cMet, TfR and integrin β1, by promoting transcript stability. (A) mRNAs were extracted from control (Ctr) and K18-depleted (K18-si) HeLa cells and qRT-PCR was performed using GAPDH as a housekeeping gene. Data are represented as mean ± S.E. from at least three independent experiments (B) Control and K18 depleted cells were left untreated or were treated with 5 μg/ml of the transcriptional inhibitor Actinomycin D for different periods of time. Transcript levels for cMet, TfR, and integrin β1 were determined by qRT-PCR. Fold changes are relative to GAPDH and were normalized to untreated control. Results are from at least three independent experiments. Statistically significant differences are indicated: *p < 0.05; **p < 0.01, ***p < 0.001, and ****p < 0.0001.