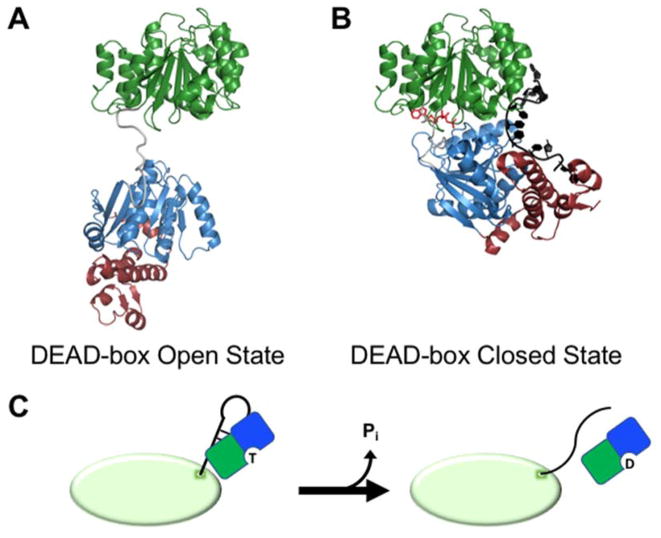
Figure 2.
Remodeling of exposed RNA duplexes by DEAD-box proteins. (A) Structural model for the open conformation of a DEAD-box protein (S. cerevisiae Mss116) prior to substrate binding (16). Conserved domains 1 and 2 are colored as in Figure 1. Mss116 and its homologs have an additional C-terminal extension (magenta) that is not present in all DEAD-box proteins. (B) Crystal structure representing the closed state of a DEAD-box protein (Mss116) following strand displacement (PDB: 3I5X) (20). The bound ssRNA strand is shown in black and the bound AMP-PNP molecule is red. (C) Model for unwinding of an exposed RNA duplex from a larger RNA or RNP by a DEAD-box helicase (shown as green and blue D1 and D2 respectively). The ATP-bound DEAD-box protein interacts with an exposed RNA helix on the surface of a structured RNA or RNP (green oval). Protein binding results in unwinding of the helix, allowing the partner strands to form new inter- or intramolecular interactions. ATP hydrolysis and release of Pi then allow the DEAD-box protein to release from the liberated ssRNA.