Figure 3.
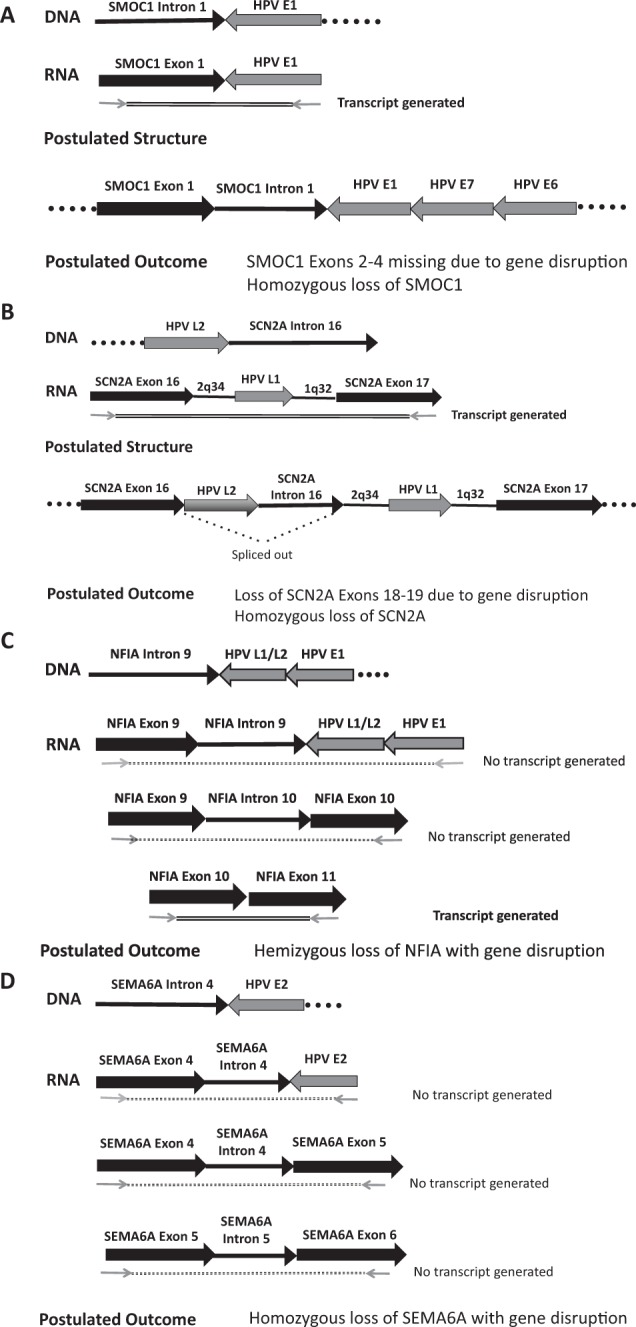
Transcript analysis of recurrent tumors that revealed gene disruption following viral integration. (A) Tumor 2049. Detection of integrated papillomavirus sequences–polymerase chain reaction (DIPS-PCR) identified a viral integration event linking human papillomavirus (HPV) E1 to SMOC1 intron 1 in the DNA. Reverse transcriptase PCR converted the RNA to complementary DNA (cDNA), and primers spanning the integration site produced a viral-host fusion transcript containing E1 and SMOC1. However, primers from SMOC1 exon 1 to exon 2 and from exon 3 to exon 4 produced no native transcripts of exon junctions spanning the site or of exons downstream of the site. We postulate that the structure includes SMOC1 exon 1 and intron 1 fused to HPV E1-E7-E6, including the viral promoter, with a loss or disruption of SMOC1 downstream exons. Since no normal transcripts were produced, we conclude that both copies of SMOC1 were lost in this tumor. (B) Tumor 0843. A fusion of HPV L2 into SCN2A intron 16 was found at the DNA level. Upon PCR testing of the cDNA using primers spanning SCN2A exon 16_exon 17, a fusion transcript containing SCN2A, HPV L1, and fragments of chromosome 2q32 and 1q32 was produced. No intact SCN2A exon-exon transcripts downstream of the integration site were produced. We postulate that HPV L2 and SCN2A intron 16 are spliced out of the structure shown above, leading to gene disruption that was accompanied by loss of the other copy of SCN2A. (C) Tumor 2238, integration event 1, NFIA intron 9 fused to HPVL1/L2 and HPVE1. This event failed to yield viral-host fusion transcripts when a primer from exon 9 was paired with an HPV E1 primer. Similarly, PCR testing using primers spanning exons 9 and exon 10 failed to produce a transcript. However, downstream exons 10 to 11 showed in-frame transcripts. It is unclear whether these transcripts come from the affected chromosome or the other copy but suggests that at least 1 copy of the gene is intact, leading us to suspect hemizygous loss of NFIA. (D) Tumor 2238, integration event 2, SEMA6A intron 4 fused to HPVE2. This integration event failed to yield viral-host fusion transcripts by PCR (SEMA6A exon 4 to E2), as well as cellular exon-exon transcripts spanning the integration site (exon 4 to exon 5). In addition, the SEMA6D-HPV E2 integration event also led to lack of cellular exon-exon transcripts downstream of the integration site (exon 5 to exon 6). Because of this, we postulate that gene disruption occurred by homozygous loss of SEMA6D.
