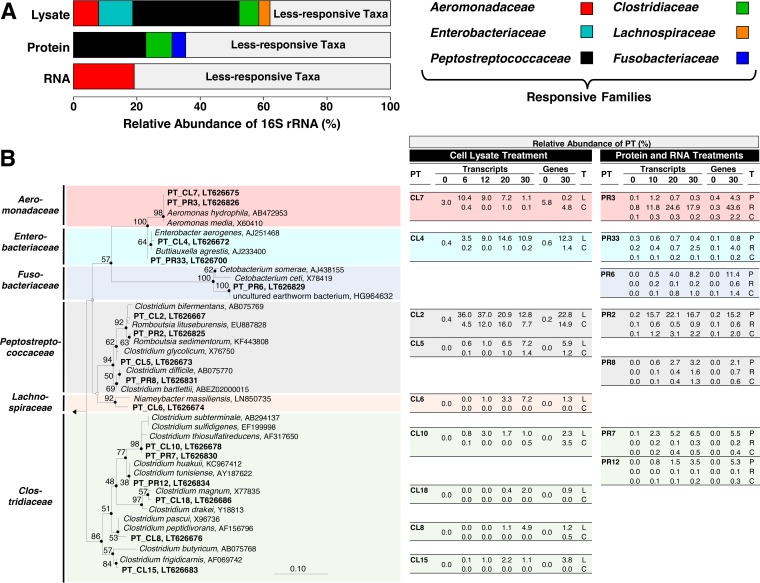
FIG 5.
Average relative abundances of 16S rRNA sequences of the most responsive families of lysate, protein, and RNA treatments (A) and 16S rRNA-based phylogenetic tree of affiliated responsive phylotypes (in bold) and reference sequences (B). (A) Families were designated most responsive when a family in a given treatment displayed a minimum increase in relative abundance of 5% above the control values in at least one of the sampling periods. The values for each family are based on the arithmetic average from all abundances detected at 6, 12, 20, and 30 h for the cell lysate treatment and at 10, 20, and 30 h for the protein or RNA treatments. (B) Phylotypes (PT) are based on a sequence similarity cutoff of 97% and were designated responsive when a phylotype in a given treatment displayed a minimum increase in relative abundance of 2% above the control values in at least one of the sampling periods. The phylotypes are derived from the analysis of either 16S rRNA (designated as transcripts) or 16S rRNA genes (designated as genes). The phylogenetic tree was calculated using the neighbor-joining, maximum parsimony, and maximum likelihood methods. Solid circles at nodes indicate congruent nodes in three trees. Empty and gray circles at nodes indicate congruent nodes in two trees (neighbor-joining congruent with maximum parsimony or maximum parsimony congruent with maximum likelihood). Branch length and bootstrap values (1,000 resamplings) are from the maximum parsimony tree. The bar indicates 0.1 changes per nucleotide. Thermotoga maritima (AE000512) was used as the outgroup. Accession numbers are shown at the end of each branch. Relative abundances (in %) of phylotypes in the table are shown for each sampling period (i.e., 0, 6, 12, 20, and 30 h for the cell lysate treatment, and 0, 10, 20, and 30 h for the protein or RNA treatments). Closely related phylotypes (i.e., >97% sequence similarity) that increased in the cell lysate (L) treatment and protein (P) or RNA (R) treatments were placed on the same horizontal level. C, unsupplemented control, T, treatment.