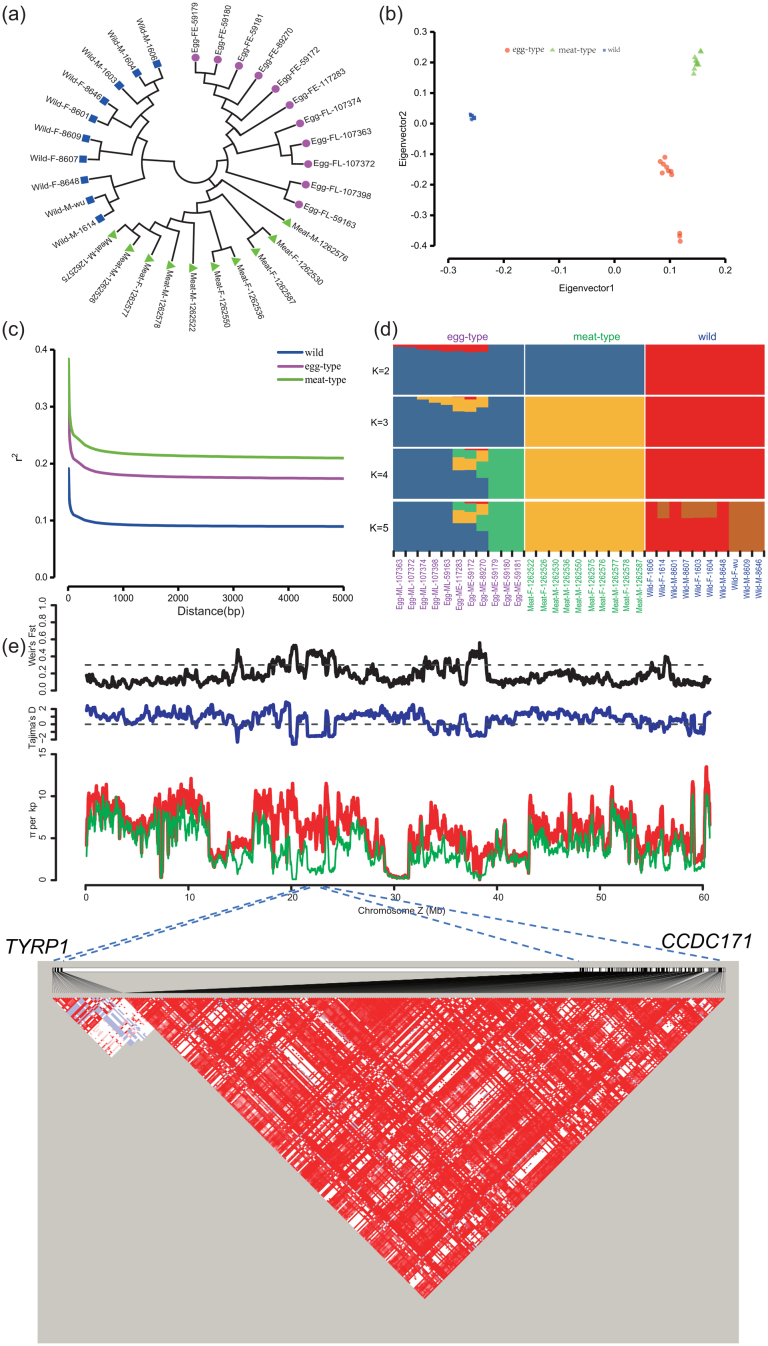
Figure 3:
Analyses of the phylogenetic relationships, population structure, LD decay, and genetic diversity between wild and domesticated quail. A) Evolutionary history was inferred using the neighbor-joining method in MEGA 6.0. B) PCA of wild quail and domesticated quail. C) LD decay curves were estimated by squared pairwise correlations of alleles against physical distance in wild quail, egg-type quail, and meat-type quail, respectively. D) Population structure analysis with the maximum likelihood score for the model K = 2. E) Nucleotide diversity between wild quail and egg-type quail across chromosome Z. Both the wild quail (red line) and the egg-type quail (green line) showed difference of diversity on chromosome Z. Plotting of Tajima's D for the egg-type group (blue line) in a 100-kb sliding window in 10-kb steps revealed the selective signal on chromosome Z. Likewise, plotting Weir's Fst (black line) on chromosome Z indicates the level of differentiation between the wild group and the egg-type group. Both CCDC171 and TYRP1 genes were located within a selective sweep region (from ∼21.5 Mb to 23.2 Mb), in which the positive signal was detected in the egg-type group. However, they exhibited a weak linkage due to the location on the different haplotype blocks.