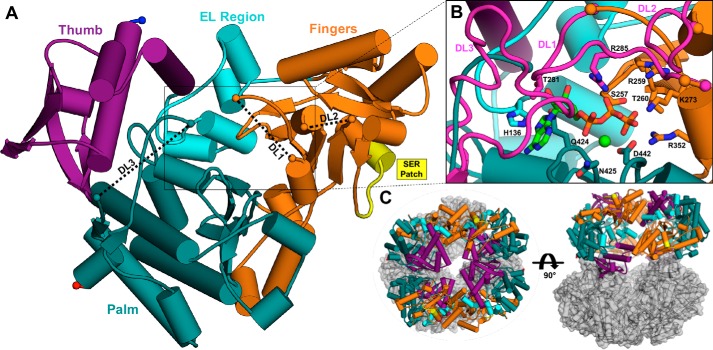
Figure 6.
The X-ray crystal structure of IucC. A, the monomeric tertiary structure of IucC surface mutant M5 colored by domain. Gaps in the model corresponding to disordered loops (DL1–3) are connected by black dashed lines for clarity. Protein chain termini are highlighted by spheres, with the N and C termini colored blue and red, respectively. The SER patch is emphasized in yellow. B, close-up view of the modeled active site. The disordered loops (DL1–3, magenta) were modeled by overlaying corresponding regions of a homology model (Fig. S4A). ATP (green carbons) and Mg2+ (light green sphere) from the crystal structure of IucA (PDB code 5JM8) are also modeled into the active site with IucC the side chains of several predicted binding residues labeled and depicted as sticks. C, the crystallographic asymmetric unit containing eight copies of IucC illustrated in two orthogonal views. Potential tetrameric (left) and dimeric (right) assemblies are highlighted using the same color scheme as panel A.