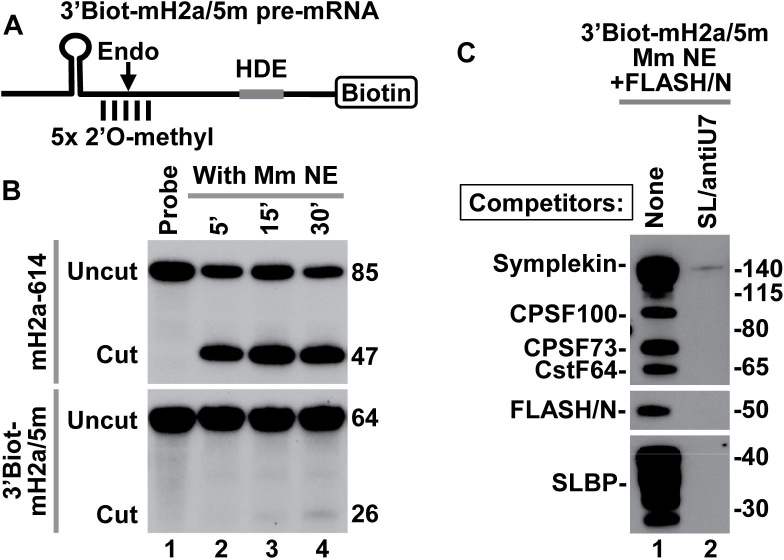
Figure 1.
3′Biot-mH2a/5m pre-mRNA is resistant to cleavage but assembles into processing complexes. (A) A schematic representation of chemically synthesized mouse-specific 3′Biot-mH2a/5m pre-mRNA (64-nt). The major cleavage site (located 5 nt downstream of the stem) and 2 nt on each side of the major cleavage site are modified with a 2′O-methyl group. Biotin is placed at the 3′ end. (B) In vitro processing of 3′Biot-mH2a/5m (bottom) and mH2a-614 (top) pre-mRNAs. mH2a-614 (85 nt) was generated by T7 transcription and contains the same HDE as 3′Biot-mH2a/5m but lacks biotin and modified nucleotides. Each pre-mRNA was labeled at the 5′ end with 32P and incubated in a mouse nuclear extract for 5, 15 and 30 min, as indicated. Probe alone is shown in lane 1. Numbers to the right indicate the length of the input pre-mRNA and the upstream cleavage product. (C) 3′Biot-mH2a/5m was incubated with a mouse myeloma nuclear extract (Mm NE) containing recombinant N-terminal FLASH (FLASH/N, amino acids 53–138) fused to GST. Assembled processing complexes were purified on streptavidin beads and analyzed by western blotting using specific antibodies (lane 1). In lane 2, formation of the processing complexes was blocked by excess SL RNA and αU7 oligonucleotide complementary to the 5′ end of mouse U7 snRNA.