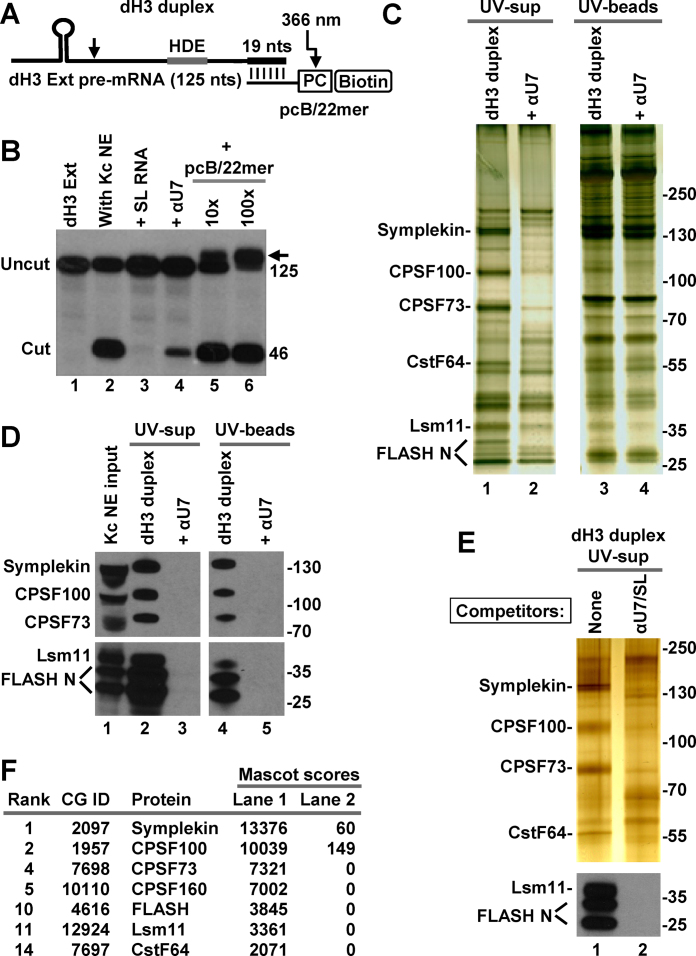
Figure 4.
Purification of Drosophila processing complexes with pre-mRNA attached to the photo-cleavable group in trans. (A) A schematic representation of the dH3 Ext duplex formed by annealing T7-generated dH3 Ext pre-mRNA and chemically synthesized 2′O-methyl pcB/22mer oligonucleotide. In pcB/22mer, biotin (B) is placed at the 5′ end and is followed by the photo-cleavable (pc) linker. The last 19 of 22 nt of pcB/22mer are complementary to the 3′ end of dH3 Ext pre-mRNA. (B) dH3 Ext pre-mRNA was labeled at the 5′ end and incubated at room temperature in a Kc nuclear extract either alone (lane 2) or in the presence of indicated oligonucleotides (lanes 3–6). SL RNA and αU7 oligonucleotide were added to a final concentration of 10 ng/μl, pcB/22mer was at either 10 ng/μl or 100 ng/μl, a 10 and 100 molar excess relative to dH3 Ext pre-mRNA, respectively. The arrow indicates an RNA duplex that survived denaturing conditions of the 7M urea gel. Lane 1 contains input dH3 Ext pre-mRNA. (C and D) dH3 Ext duplex was incubated with a Drosophila Kc nuclear extract to form processing complexes (lanes 1 and 3 in panel C, and lanes 2 and 4 in panel D). As a negative control, formation of the processing complexes was blocked by αU7 oligonucleotide complementary to the 5′ end of Drosophila U7 snRNA (lanes 2 and 4 in panel C, and lanes 3 and 5 in panel D). Proteins bound to the duplex RNA were purified on streptavidin beads and UV-eluted. A fraction of the UV-eluted material (UV-sups, lanes 1 and 2) and the beads following UV-elution (UV-beads, lanes 3 and 4) was analyzed by silver staining (panel C) or western blotting (panel D). (E and F) Drosophila processing complexes were assembled on the duplex RNA and purified, as described above with the difference that negative control contained both SL RNA and αU7 oligonucleotide. A fraction of the UV-eluted samples was analyzed by silver staining and western blotting for selected proteins (panel E). The remainder was concentrated by precipitation with acetone, subjected to a brief electrophoresis sufficient for proteins to enter the gel, in-gel digested with trypsin and analyzed by mass spectrometry (panel F). Only top scoring processing factors are listed.