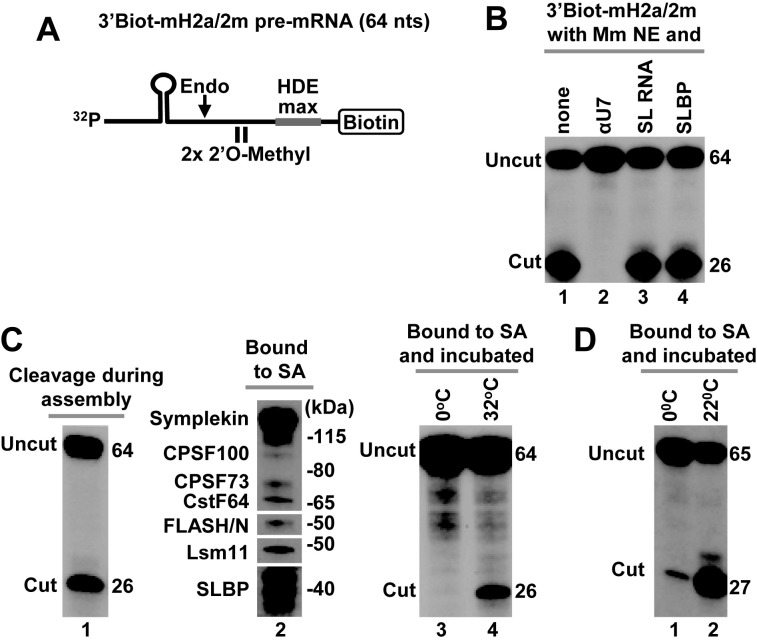
Figure 5.
Cleavage activity of immobilized mouse and Drosophila processing complexes. (A) A schematic representation of chemically synthesized mouse-specific 3′Biot-mH2a/2m pre-mRNA (64 nt). The two 2′O-methyl groups placed immediately upstream of the HDE have no effect on endonucleolytic cleavage but prevent subsequent 5′-3′ degradation of the downstream cleavage product. Biotin is placed at the 3′ end and the HDE is modified by two point mutations to maximize its base pair interaction with U7 snRNA (HDE max). (B) In vitro processing of 3′Biot-mH2a/2m pre-mRNA in a mouse nuclear extract alone (lane 1) or containing indicated reagents: αU7 oligonucleotide (10 ng/μl), SL RNA (10 ng/μl) or human baculovirus-expressed SLBP (25 ng/μl). (C) Mouse processing complexes (lanes 1–4) were assembled on ice by incubating 32P-labeled 3′Biot-mH2a/2m pre-mRNA with a mouse nuclear extract containing recombinant N-terminal FLASH. During this step, significant cleavage occurred reducing the amount of fully assembled processing complexes (lane 1). Length in nucleotides of the input 3′Biot-mH2a/2m pre-mRNA and of the upstream cleavage product in nucleotides is shown to the right. The assembled complexes were immobilized on streptavidin beads, thoroughly washed and analyzed by western blotting for the presence of SLBP and selected subunits of the U7 snRNP (lane 2) or incubated with a gentle agitation at indicated temperatures to determine their ability to support cleavage (lanes 3 and 4). As explained above, in mouse nuclear extracts SLBP is partially degraded and in SDS gels migrates as a group of closely spaced bands rather than a single full length species of ∼45 kDa. (D) Drosophila processing complexes were assembled on Drosophila-specific 3′Biot-dH3/2m pre-mRNA. The complexes were immobilized on streptavidin beads, washed and incubated with a gentle agitation at 0°C (lane 1) or 22°C (lane 2) to determine their ability to support cleavage.