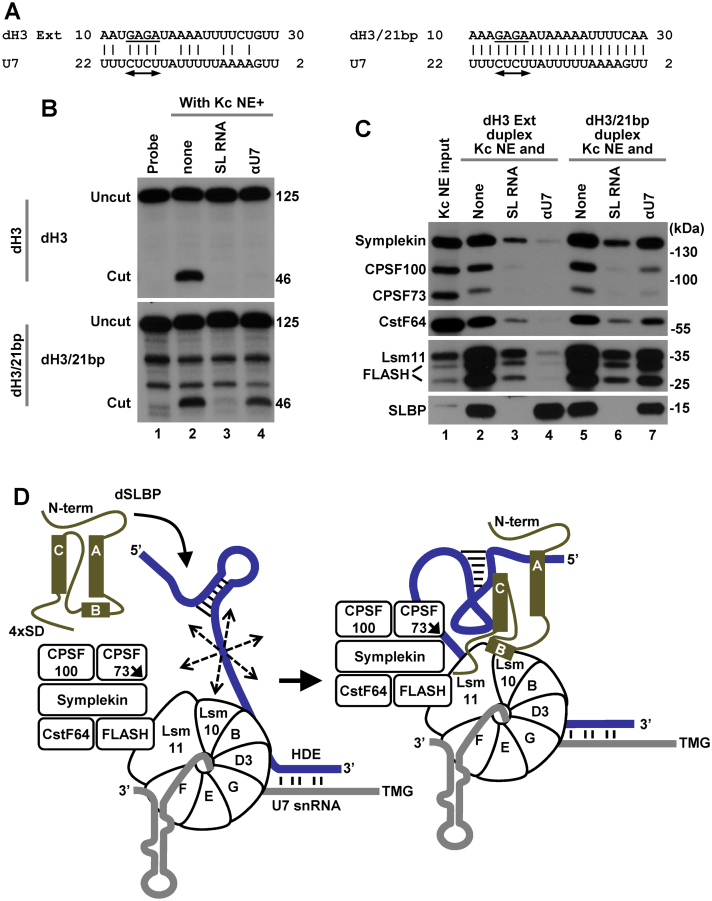
Figure 7.
Effects of increasing the base pairing potential between histone pre-mRNA and Drosophila U7 snRNA. (A) A proposed base pair alignment between the HDE of dH3 Ext (left) and dH3/21bp (right) pre-mRNAs and Drosophila U7 snRNA. The HDE starts 10 nt 3′ of the cleavage site and contains the GAGA sequence (underlined) that is essential for processing and is predicted to form an obligatory duplex with the UCUC motif (indicated with a double-headed arrow) conserved in most U7 snRNAs. (B) Processing of 5′-labeled dH3 Ext (top) and dH3/21bp (bottom) pre-mRNAs in a Kc nuclear extract either lacking (lane 2) or containing indicated competitors (lanes 3 and 4). The input RNA is shown in lane 1. Length of the uncut pre-mRNAs and the upstream cleavage products (cut) in nucleotides is shown to the right (C). dH3 Ext and dH3/21bp pre-mRNAs were annealed to pcB/21mer and the resultant duplexes incubated with a Kc nuclear extract in the absence or presence of oligonucleotide competitors, as indicated. The assembled processing complexes were immobilized on streptavidin beads and analyzed by western blotting for the presence of SLBP and major subunits of the U7 snRNP. (D) A hypothetical model explaining essential role of Drosophila in processing. The three α helices of the RBD of Drosophila SLBP are depicted as rectangles and the repeated SD motif is shown at the C-terminal region. In the absence of Drosophila SLBP, U7 snRNP inefficiently binds histone pre-mRNA (blue line) via limited base paring between U7 snRNA (gray line) and the HDE. The bound U7 snRNP in spite of containing all necessary subunits is unable to carry out the cleavage reaction, possibly due to the excessive flexibility of the pre-mRNA substrate (illustrated by multiple arrows). SLBP bound to the stem-loop uses helix B and the C-terminal region to promote the recruitment of the U7 snRNP to the HDE, likely by contacting Lsm11 bound to FLASH and possibly Lsm10. The same network of interactions may result in a major structural rearrangement of the processing complex and stable alignment of the active site of CPSF73 (indicated with an arrow) with the pre-mRNA.