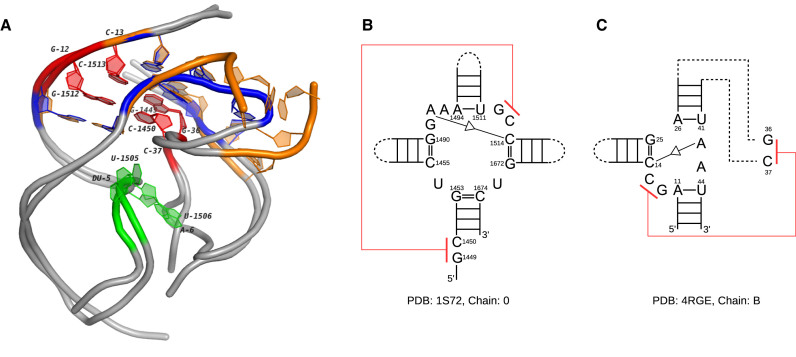
Figure 9.
The 3D and secondary structures of two multi-loops in a 23S rRNA and an env22 twister ribozyme. (A) The 3D docking of two loops. The orange tube represents the multi-loop in the 23S rRNA (PDB ID: 1S72, chain: 0) and the blue tube represents the multi-loop in the twister ribozyme (PDB ID: 4RGE, chain: B). The extended regions of both loops are shown in gray. The nucleotides involved in the pseudoknots are labeled (G1512-C1450 and C1513-G1449 in 1S72, G12-C37 and C13-G36 in 4RGE). (B) The secondary structure of the multi-loop in the 23S rRNA. (C) The secondary structure of the multi-loop in the twister ribozyme. The pseudoknots in (B) and (C) are marked in red. The self-cleavage sites in 4RGE and the corresponding nucleotides in 1S72 are marked in green.