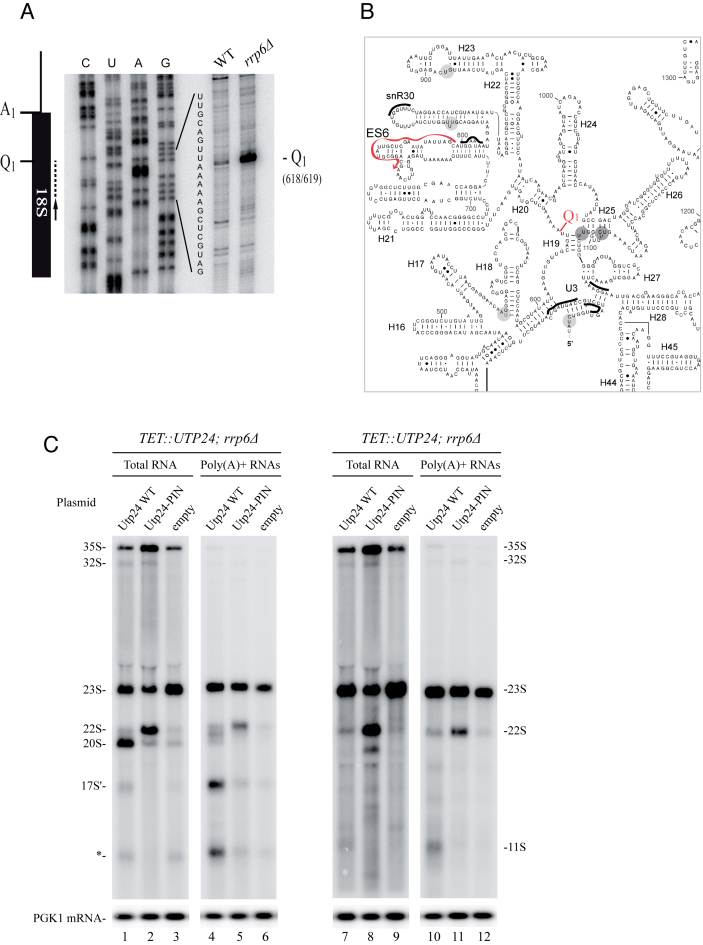
Figure 7.
23S pre-rRNA is cleaved at Q1 site and the PIN domain of Utp24 is required for efficient cleavage. (A) RNAs extracted from WT or rrp6Δ strains were analyzed by primer extension using probe 1510 (schematized by the arrow). A PCR fragment containing 18S sequence was used to generate a sequencing ladder leading to the identification of Q1 site. (B) Predicted secondary structure of the 18S rRNA central domain in Saccharomyces cerevisiae. Utp24 crosslinking sites are marked on the sequence and shades indicate peak height with the highest peak shown in dark grey. Q1 site (18S 618/619) is indicated. Oligonucleotide used to map Q1 site is annotated (red arrow). (C) Northern analysis of pre-rRNA processing in the pTET::utp24–3HA strain transformed with a plasmid expressing either WT Utp24 or the PIN mutant (D68N) his-tagged Utp24 protein, or an empty vector. RNAs were isolated from mid log phase cells grown 8 h in presence of doxycycline to deplete endogenous Utp24 protein. Aliquots were collected and total RNAs were extracted and subjected to poly(A)+ affinity purification on oligo-dT-coated beads. Total RNAs (lanes 1–3 and 7–9) and poly(A)+ RNAs (lanes 4–6 and 10–12) were separated on an 1.2% agarose gel and detected by northern hybridization with specific oligonucleotide probes. Left panels (lanes 1–6) were hybridized with ITS1 probe (004), right panels (lanes 7–12) were hybridized with ETS1 probe (1699). Loading was assessed by hybridizing the PGK1 mRNA (lower panel) with probe 403.