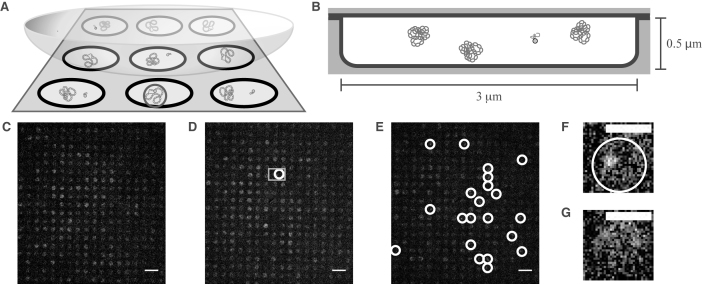
Figure 2.
CLiC nanopit microscopy and binding events. (A) Schematic of the CLiC method confining plasmids and labeled oligo-probes to pits made using micro/nano lithography techniques (28). The circle in the front-center pit demonstrates a pit containing a bound probe–plasmid complex. (B) Schematic of plasmids and a freely diffusing probe confined to a circular pit of 3-μm diameter and 0.5-μm depth. Each pit contains ∼24 plasmids and a single labeled probe. (C) Fluorescence image of mixture of probes and pUC19 plasmids with mean superhelical density <σ> = 0 and standard deviation sσ = 0.03 confined to pits. ( D) Fluorescence image of mixture of probes and pUC19 plasmids with superhelical density <σ> = − 0.050, sσ = 0.07 confined to pits. The gray square denotes the two pits containing single probes in F and G. (E) Fluorescence image of mixture of probes and pUC19 plasmids with <σ> = − 0.127, sσ = 0.06 confined to pits. White circles show binding events. (F) Pit containing a bound probe–plasmid complex from the data set shown in D. (G) Pit containing a freely diffusing probe from the data set shown in D. Scale bars in C–E denote 10 μm, while those in F and G are 3 μm. Images in C–G had their background subtracted using a rolling ball algorithm with a radius of 50 pixels, and a Gaussian-blur filter with radius 0.5 pixels was applied to each.