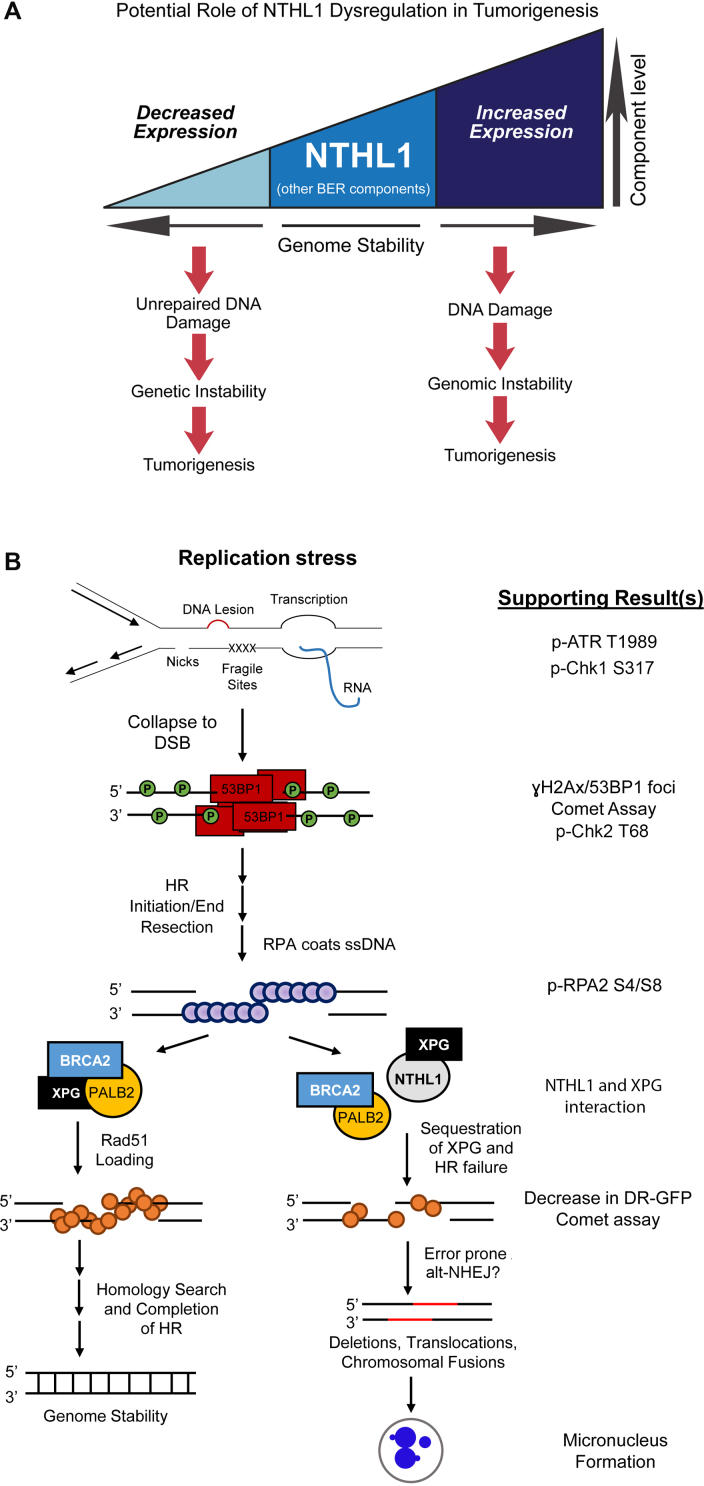
Figure 7.
Proposed models for NTHL1 regulation/dysregulation. (A) Loss or decreased expression of NTHL1 is linked to a novel cancer predisposition syndrome (13,14), presumably through lack of DNA repair for NTHL1 substrates, yielding mutations, genetic instability, and ultimately tumorigenesis. Alternatively, increased expression of NTHL1 protein levels can lead to DNA damage that drives genomic instability and contributes to tumorigenesis. Therefore, optimal protein levels of BER components are needed to balance the cellular need for DNA damage repair without initiating genomic instability. (B) A model for events leading to genomic instability. Results supporting the model that is presented in this work are listed on the right. Replication stress that arises during S phase may occur through multiple means, including unrepaired DNA lesions, accumulated DNA repair intermediates, nicks in the phosphodiester backbone, fragile sites, and encounters with transcription, among other causes. A collapsed replication fork results in a DSB that is repaired by homologous recombination (HR). Phosphorylation of γH2Ax and binding of 53BP1 signals the presence of a DSB. BRCA1 displaces 53BP1 in S phase to promote end resection to initiate HR. Single-stranded DNA is then coated by RPA, and RPA phosphorylation on serine 4 and serine 8 (pRPA S4/S8) signal for RAD51 loading. Initiation of HR requires formation of the RAD51 presynaptic filament through displacement of RPA by BRCA2 with PALB2 and DSS1, and efficient RAD51 loading is also dependent on XPG. Completed HR results in genome stability, as a homologous template is used to repair the DSB. Upon NTHL1 overexpression, XPG could be sequestered through protein interaction, thus inhibiting functions required for proper RAD51 loading. This could result in chronic replication stress signaling, as evidenced by pATR T1989. Inefficient RAD51 loading onto DNA results in HR failure, and competing end joining increases. This end joining may include alt-NHEJ to salvage DSB repair, as DNA resection has already occurred. Alt-NHEJ is error prone and can lead to genome instability, as assessed by micronucleus formation.