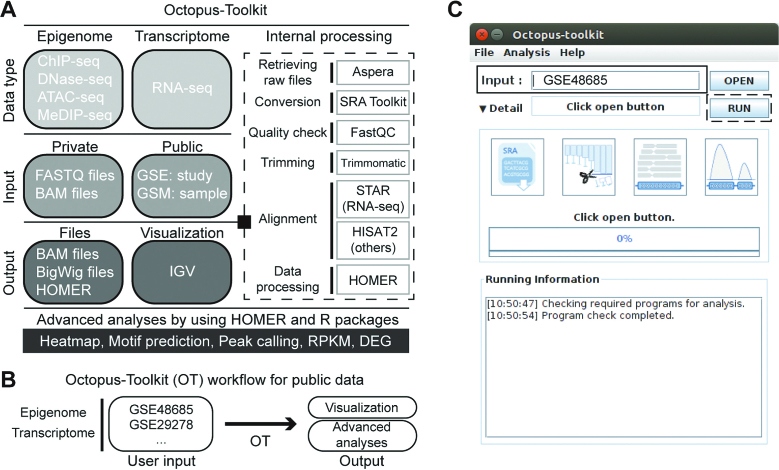
Figure 1.
Octopus-Toolkit workflow. (A) Detailed information on data types, input, and output for Octopus-Toolkit is shown. Programs associated with Octopus-Toolkit and their purposes are described (dashed line box). (B) An example of its use is depicted. (C) Graphical user interface of Octopus-Toolkit is shown. The only input required for Octopus-Toolkit is an accession number for epigenomic and transcriptomic NGS data sets (GSE accession number) or a single piece of NGS data (GSM accession number) (black box). Multiple NGS data sets can be sequentially processed by providing a list of GSE (or GSM) accession numbers as a text file. Octopus-Toolkit runs all the steps after the Run icon (dotted line box) is clicked on.