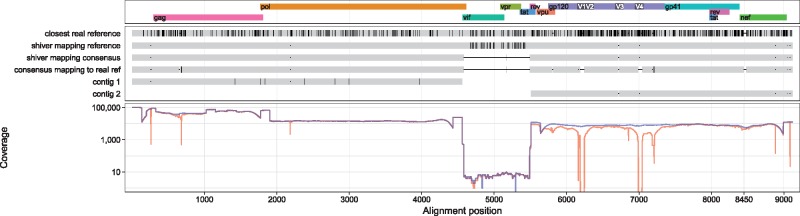
Figure 5.
Top panel: HIV genes in their reading frames. Middle panel: sequences for the Miseq sample ERR732065. From top to bottom these are the closest identified real reference (see main text), the reference created and used for mapping by shiver, the consensus of reads mapped to shiver reference, the consensus of the same reads mapped to the real reference, and the contigs generated by de novo assembly. Vertical black lines inside sequences in the alignment denote single nucleotide polymorphisms (SNPs), defined here relative to the most common base among these sequences. Horizontal black lines indicate a lack of bases, i.e. a deletion relative to another sequence in the alignment or, for the two consensuses, simply missing sequence due to insufficient coverage. Bottom panel: the coverage (number of mapped reads) for the shiver reference in blue, and for the real reference in red. Mapping problems at Position 8450 are shown in detail in Fig. 2. Where the real reference and the sample differ by many close SNPs or an indel, differences often arise between the shiver consensus and the consensuses mapping to the real reference. The coverage plot beneath the sequences shows that at such points, the coverage mapping to the real reference almost always drops below the coverage mapping to the shiver reference; given that the same reads are being mapped to the same part of the genome with the same mapping parameters, this strongly suggests that the shiver consensus is more accurate. This is the case at Position 8450 in this figure, in the nef gene; the problem mapping to the real reference here was shown in detail in Fig. 2. Though the coverage here drops due to the problem aligning the reads, it is still more than 4,000, showing that a large absolute number of reads is no guarantee of accuracy. Mapping to the shiver reference on the other hand, coverage remains locally smooth. Similar errors mapping to the real reference in this figure can be seen in gag and in five different places in gp120.