Figure 1.
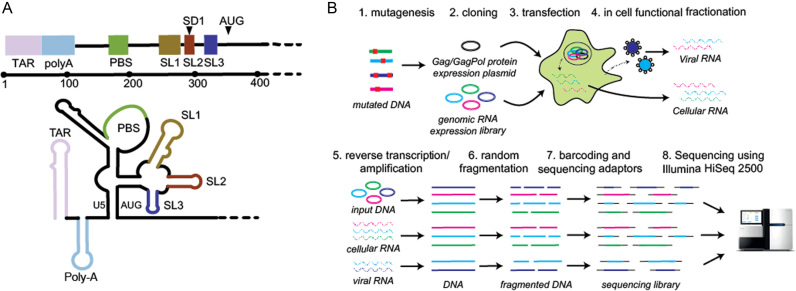
(A) The HIV-1 5′UTR folds into a series of structural domains that control key steps of the HIV-1 life cycle including transcription, translation, export, packaging and reverse transcription. From 5′ to 3′ these structural domains are: transactivation response (TAR) for transcription; PolyA stem loop for polyadenylation; the primer binding site (PBS) for reverse transcription; SL1 promotes gRNA dimerization; SL2 contains the major splice donor (SD) site; SL3 has historically been considered the major packaging signal (Psi); the sequences surrounding the AUG start codon are thought to be involved in a base-pairing interaction with the upstream U5 region. (B) In cell Mutational Interference Mapping Experiment (in cell MIME). The proviral genome is randomly mutated using error prone PCR, and subsequently cloned into a gRNA expression vector. The structural and enzymatic proteins, Gag and Gag-Pol are expressed from a separate expression plasmid. Co-transfection of the mutant library and Gag/Gag-Pol expression plasmid into 293T cells leads to the transcription of mutant RNAs and subsequent sorting of functional and non-functional RNA populations by the viral and cellular machinery. Viral RNA present in cells and virus is reverse transcribed. Viral cDNA and the input DNA plasmid is amplified, fragmented, barcoded, sequenced on an Illumina HiSeq2500, and analysed using the MIMEAnTo software.