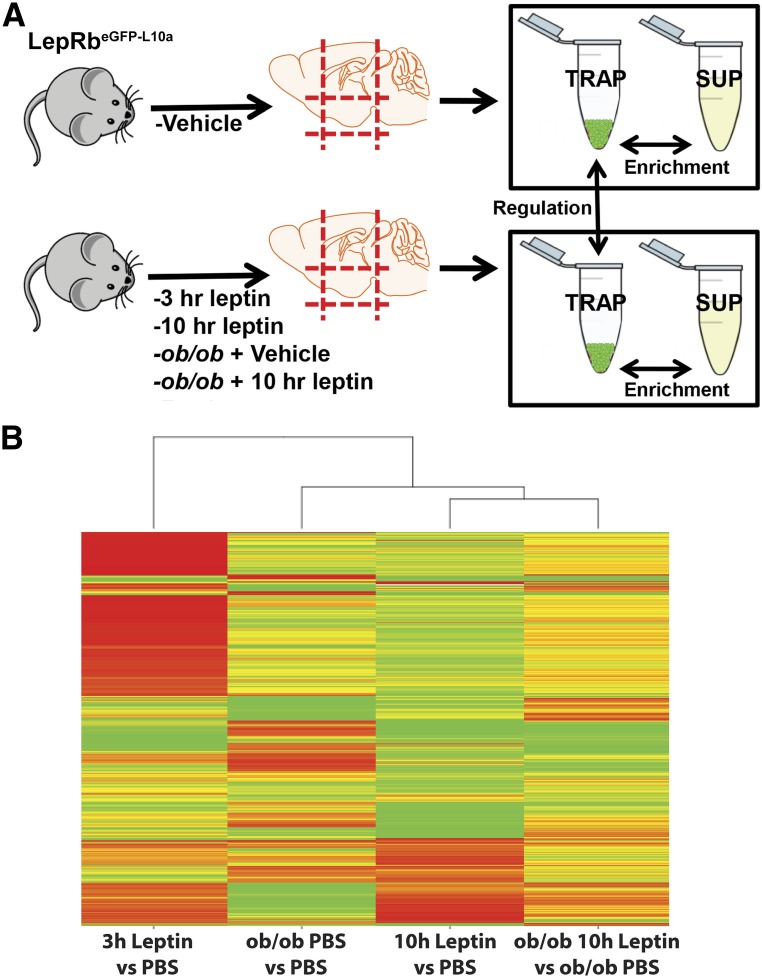
Figure 1.
TRAP-seq analysis to identify leptin-regulated genes in hypothalamic LepRb neurons. A: Experimental design: LepRbeGFP-L10a mice were subjected to a variety of perturbations in leptin levels before hypothalami were dissected and mRNA-containing ribosomes from LepRb neurons were TRAP purified. TRAP and supernatant mRNAs were subjected to RNA-seq; comparisons of TRAP and supernatant fractions revealed the identity of genes enriched in LepRb neurons under each condition, and comparing TRAP material among conditions revealed the regulation of these enriched genes by leptin. B: A heat map demonstrating changes in gene expression in LepRb neuron-derived TRAP samples across conditions. Fold change scale is from 4.1-fold increased (green) to –9.7-fold decreased (red). Euclidean distance among the conditions (maximum 50) is shown along the top. hr, hour; SUP, supernatant.