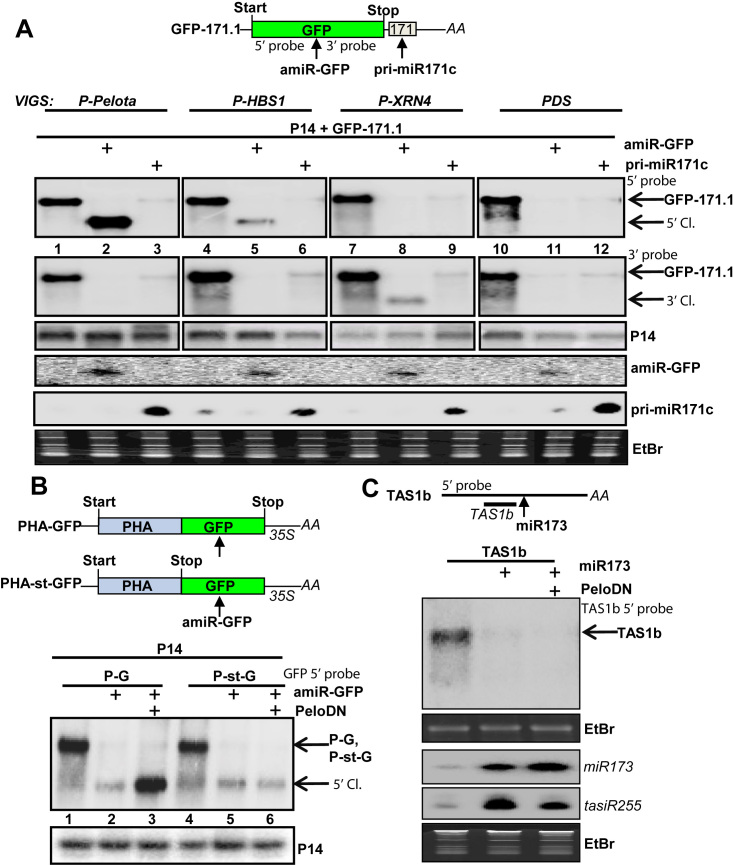
Figure 3.
NSD eliminates the 5′ cleavage fragments of miRISC when the cleavage occurs in the coding region. (A) The role of NSD components and XRN4 in the degradation of miRISC-generated cleavage fragments. PDS-, P-XRN4-, P-Pelota- and P-HBS1-silenced plants were co-agroinfiltrated with P14 internal control and GFP-171.1 miRNA sensor or P14 and GFP-171.1 constructs were co-expressed with either amiR-GFP or pri-miR171c constructs. amiR-GFP targets the sensor mRNA within the coding region, while pri-miR171c targets the sensor within the 3′UTR. Note that the weak miR171 signals in the samples without the agroinfiltrated pri-miR171c originates from the endogenous locus. (B) The effect of PeloDN on the accumulation of 5′ cleavage fragments of amiR-GFP-programmed RISC. PHA-GFP fusion (P-G) construct and PHA-st-GFP (P-st-G), a similar sensor construct in which a stop codon is present between the PHA and GFP reporter genes, were co-agroinfiltrated with amiR-GFP, or with amir-GFP and a dominant-negative version of Arabidopsis Pelota1 (PeloDN) (p14 was used as internal control). 5′ cleavage product accumulates only when PeloDN is present and the amir-GFP–directed cleveage occurs within the coding region. (C) NSD does not affect the tasiRNA silencing pathway. N. benthamiana leaves were co-agroinfiltrated with TAS1b mRNA and miR173 expressing constructs, or with TAS1b, miR173 and PeloDN expressing constructs. RNA gel blot assays were conducted to monitor the abundance of miR173, TAS1b and the tasiR255 RNAs. TAS1b is cleaved by miR173-programmed RISC, and then tasiR255 is generated by RDR6 and DCL4 from the 3′ cleavage product. The presence of PeloDN does not stabilize the miR173-generated TAS1b cleavage product. As P14 modifies the tasiRNA pathway, P14 was not co-expressed.