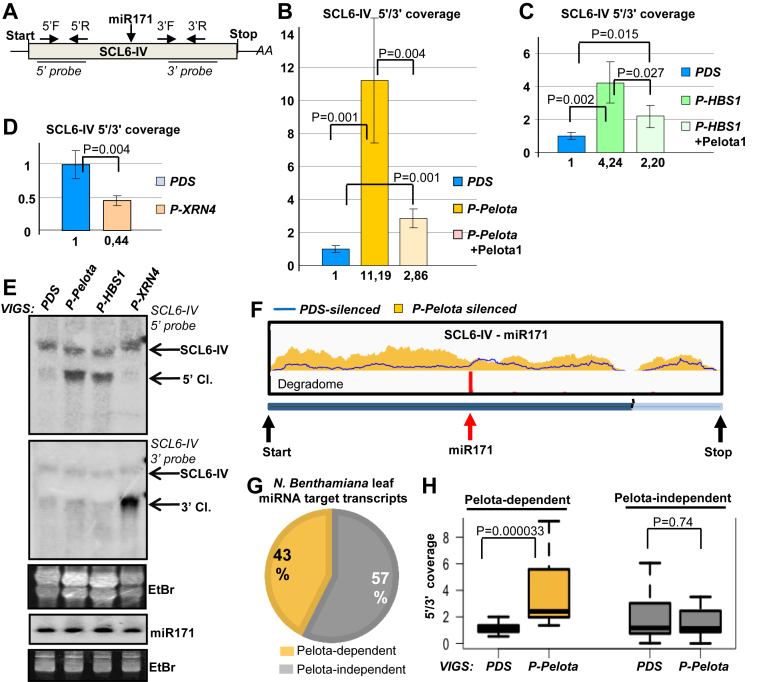
Figure 4.
NSD is involved in the degradation of 5′ cleavage fragments of several endogenous miRNA targets. (A) The schematic, non-proportional representation of the N. benthamiana SCL6-IV miRNA171 target transcript. The primer pairs that were used to measure the accumulation of the 5′ (5′F and 5′R) and 3′ (3′F and 3′R) cleavage fragments and the probes that were used for the northern assay are shown. (B and C) The 5′ cleavage fragments of SCL6-IV miR171 target mRNAs are overaccumulated in NSD deficient leaves. qRT-PCR assays were carried out from PDS-, P-Pelota- and P-HBS1-silenced leaves, and from Pelota1-complemented P-Pelota- and P-HBS1-silenced leaves. (D) qRT-PCR experiments show that the 3′ cleavage fragments of SCL6-IV mRNAs are overaccumulated in P-XRN4-silenced leaves. (E) RNA gel blot assay was carried out to study the accumulation of the 5′ and the 3′ cleavage products (5′ Cl. and 3′ Cl.) of SCL6-IV mRNA in PDS-, P-XRN4-, P-Pelota- and P-HBS1-silenced leaves. Note that VIGS did not alter the expression of the miRNA171. (F–H) Comparative RNA-seq assay conducted from PDS- and P-Pelota-silenced plants show that Pelota plays a role in the degradation of a subset of endogenous miRNA target transcripts. (F) The coverage of the SCL6-IV transcript. Yellow columns show the number of reads at a given position from P-Pelota, while blue line indicates number of reads from PDS-silenced control. The degradome peak (red column) and the miRNA cleavage site (red arrow) are marked. (G andH) The 5′/3′coverage ratio (see main text) of miRISC targets from PDS- and P-Pelota silenced plants were compared. Transcript whose 5′/3′ coverage ratio was at least 1.5-fold higher in the P-Pelota–silenced plants were defined as Pelota-dependent.