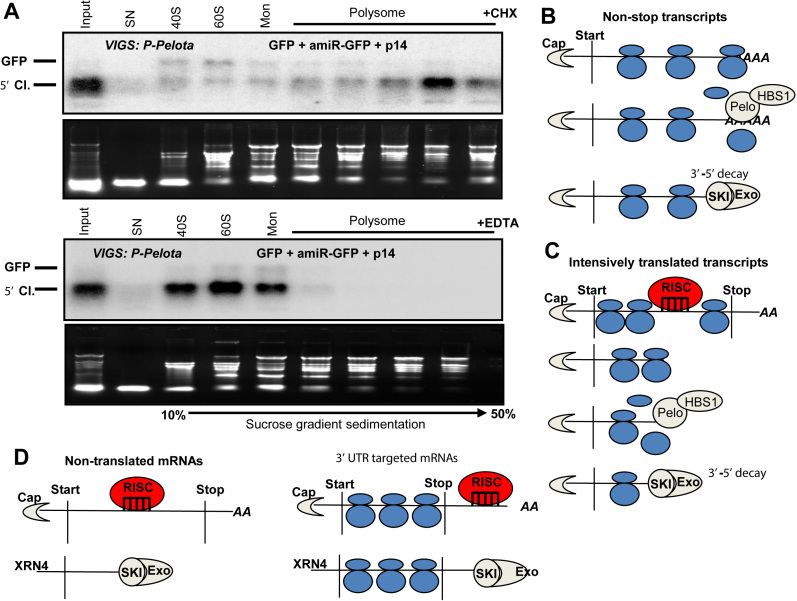
Figure 6.
Model of plant NSD. (A) In NSD-deficient cells, the 5′ cleavage fragments of miRISC are associated with ribosomes. Extracts were isolated with cycloheximide (CHX) or EDTA containing buffer (upper and bottom panel) from P-Pelota-silenced (P-Pelota) leaves, in which GFP, amiR-GFP and P14 were co-expressed. These extracts were subjected to sucrose gradient sedimentation, and then the fractions were analyzed by RNA gel blot assays. The blots were hybridized with GFP 5′ fragment probe. Note that the 5′ cleavage fragment (5′ Cl.) accumulates in the polysomal fractions in the CHX containing samples, while polysomal migration of 5′ fragment is disrupted by the addition of EDTA. Mon indicates monosomal fraction. (B) The role of NSD in the decay of nonstop and (C) RISC cleavage generated stop codon-less transcripts. It is not known whether SKI-exosome mediated decay occurs when the mRNA is still associated with ribosomes or SKI-exosome degrades mRNAs, from which the ribosomes have already removed. (D) NSD is not required for the 5′ fragment decay when si/miRISC cuts outside of the coding region or if the target transcript is not translated. For details, see the main text.