Figure 5. FIGURE 5: Adaptive Cyc8 mutations are specific to the TPR9 and TPR10 motifs.
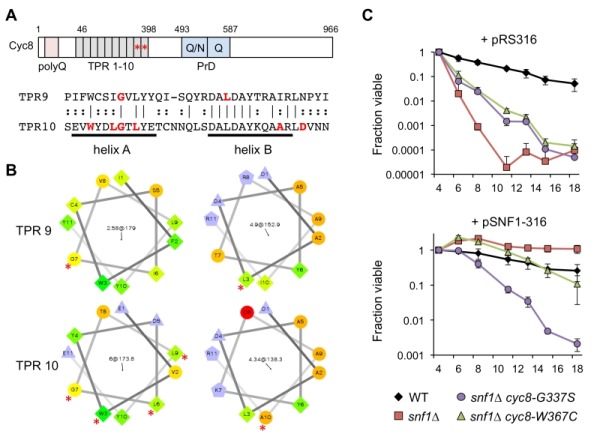
(A) Domain organization of the Cyc8 protein, indicating an N-terminal poly-glutamine (polyQ) tract, an array of 10 TPR motifs from amino acids 46 to 398, and the prion domain (PrD) consisting of polyQ/N and polyQ repeats. Asterisks indicate the location of mutations in the TPR9 and TPR10 motifs. An alignment of the TPR9 and TPR10 motifs is shown, indicating amino acids that were changed in the adapted strains. Note the mutations are distributed across the characteristic amphipathic α-helices.
(B) Helical wheel representation of the A and B α-helices of TPR9 and TPR10 generated online (http://rzlab.ucr.edu/scripts/wheel/wheel.cgi . Mutated residues are indicated by an asterisk.
(C) Quantitative CLS when SNF1 is restored to a cyc8 TPR9 mutant (JS1416, snf1∆ cyc8-G337S) or a cyc8 TPR10 mutant (JS1417, snf1∆ cyc8-W367C) by transformation with a CEN-ARS URA3 plasmid containing the SNF1 gene (pSNF1-316). The empty URA3 vector (pRS316) was also transformed as a control. Cells were maintained in SC-ura medium with 2% glucose throughout the experiment. Error bars indicate standard deviations.
