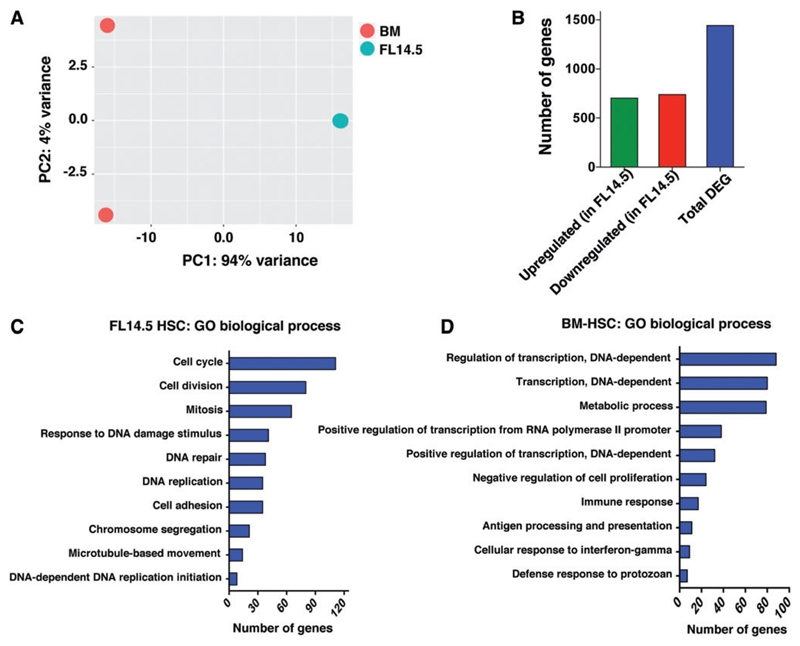
Fig. 1. Genome-scale transcriptional profiling between murine FL and ABM LT-HSC.
(A) RNASeq analysis was performed as described in an earlier publication [23]. Principle component analysis across different cell populations. (B) Number of differentially regulated genes obtained after pair-wise comparison between LT-HSCs from murine FL E14.5 and ABM. Threshold used is: Log2FC ≥1 false discovery rate ≤0.05. (C, D) Gene Set Enrichment Analysis for GO biological process for genes differentially expressed between murine FL E14.5 and ABM HSCs using the web-based application Genecodis3. ABM, adult bone marrow; FL, fetal liver; GO, gene ontology; LT-HSC, long-term hematopoietic stem cell. Color images available online at www.liebertpub.com/scd