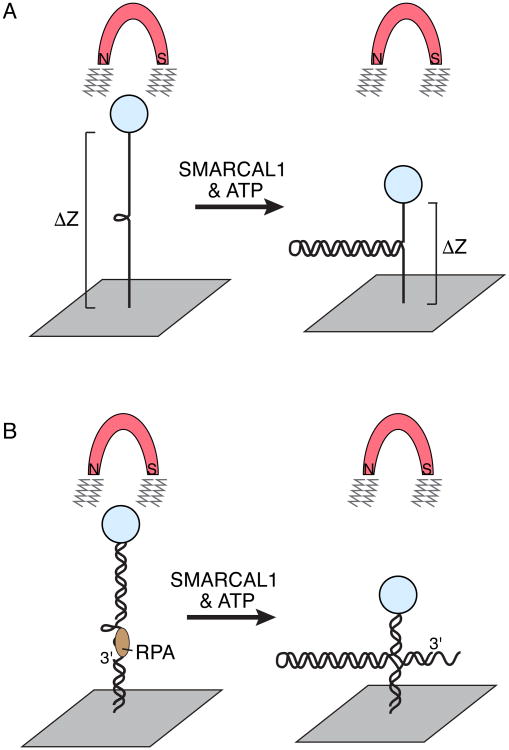
Figure 3.
Single molecule magnetic tweezer experiment to monitor SMARCAL1 annealing and fork reversal activities. (A) A 1.2kb DNA hairpin substrate is attached to a glass slide on one end and a magnetic bead on the other. Application of a magnetic field will stretch the DNA and unwind the duplex except for the last 20-30 base pairs because of their high GC content. MARCAL1 catalyzes re-annealing of the duplex DNA against the applied force, which is measured as a change in the distance of the bead from the glass slide. This experimental setup revealed that a single molecule of SMARCAL1 catalyzes bursts of repetitive annealing. (B) Addition of oligonucleotides to the stretched DNA allows the creation of substrates that mimic replication forks with ssDNA gaps on either the leading strand (depicted) or lagging template strands. RPA can be added to bind the ssDNA. This experimental set up revealed that RPA increases the distance SMARCAL1 moves per annealing event when it is bound to the leading template strand.