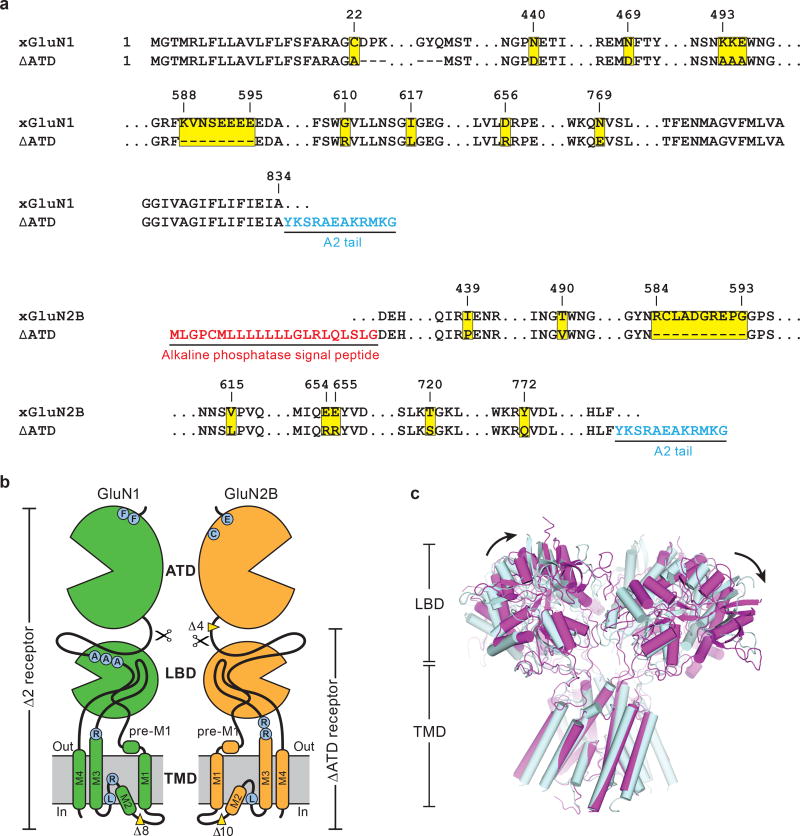
Extended Data Figure 1. The ∆ATD NMDA receptor construct and structure.
a, Selected amino acid sequences of constructs used in these studies are compared to the wild-type sequence to highlight mutations in both subunits. Locations of mutated sites and deletions are highlighted in yellow squares. Insertions of sequence are in blue or red. The ‘A2 tail’ is derived from residues 837-847 of GluA2 AMPA receptor carboxy terminus (NP_058957). b, Cartoon representation shows the GluN1 and GluN2B subunit constructs and modifications of the ∆ATD receptor. The locations of point mutations are highlighted in blue circles and the deletions are defined by yellow wedges. c, Superposition of the two ∆ATD NMDA receptors in the crystallographic asymmetric unit, aligned by the TMD. Black arrows show the shift between receptor 1 (light blue) and receptor 2 (magenta).