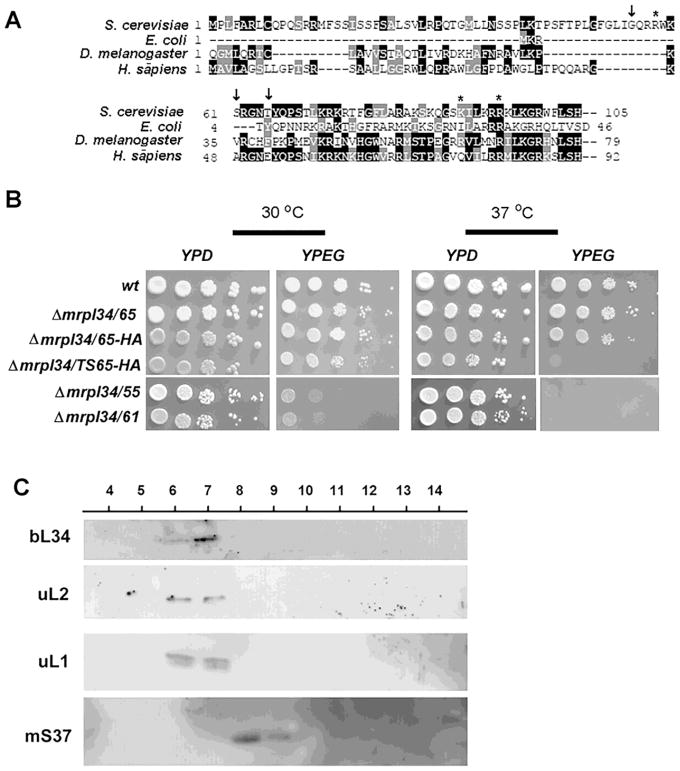
Figure 5. Expression, growth and localization of bL34 truncated variants.
(A) Alignment of the N-terminal end of L34 sequences from H. sapiens, S. cerevisiae, E. coli and A. thaliana. The arrows (i) indicate in the Saccharomyces cerevisiae sequence the position of the N-terminal fusions with “HA” tags, and asterisks indicate the position of mutations present in the mrpl34/ts mutants. (B) Growth properties of truncated variants of bL34 starting with T65 (mrpl34/65) G55 (mrpl34/55) and S61 (mrpl34/61) as well as truncated T65 containing the HA fusion (mrpl34/65-HA) and truncated T65 construction obtained from ts2 allele also fused to HA (mrpl34/TS65-HA). Cultured cells were serially diluted and spotted in rich glucose media (YPD) and rich ethanol-glycerol media (YPEG) plates. Plates were incubated at 30° C and 37° C as indicated and photographed after two days of incubation. (C) Mitochondrial ribosomes were purified by sedimentation through a sucrose cushion as described in the Material and Methods section. 20 mL of each fraction was separated by SDS-PAGE in a 12% polyacrylamide gel. Proteins were transferred to nitrocellulose and reacted with rabbit polyclonal antibodies against the large subunits uL1p, uL2p and the small subunit protein mS37. Proteins were visualized with Super Signal (Pierce Biochemicals) following a secondary reaction with peroxidase-coupled anti-rabbit antibody.