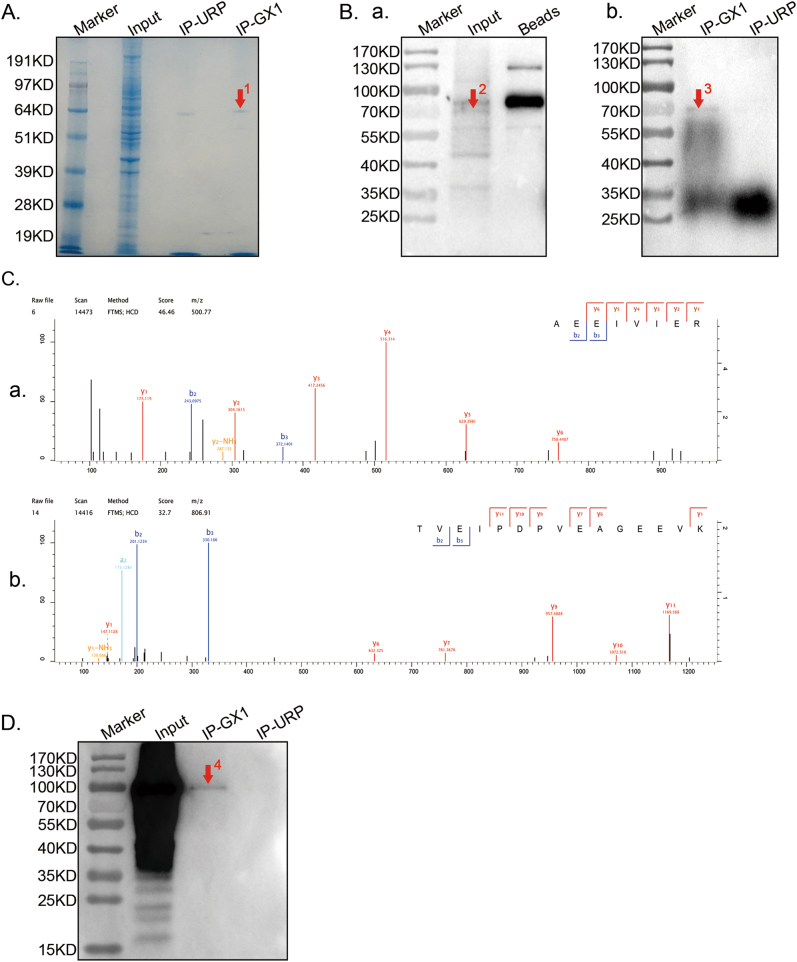
Fig. 2. Screening the receptor of GX1 in co-HUVECs.
a, b Proteins that bound to GX1 were enriched from co-HUVEC lysates by co-immunoprecipitation, separated by SDS-PAGE, subjected to Coomassie blue staining (a) and incubated with biotin-GX1(b). One immune-reactive band was detected at approximately 70 kDa in both the input (indicated by red arrow 2) and IP-GX1 lane (protein binding to GX1, indicated by red arrows 1 and 3). The IP-URP (protein binding to URP) and Beads (protein binding to magnetic beads) lanes were used as negative controls. c (panels a and b) TGM2 was identified by Q-Exactive mass spectrometry. The MS/MS spectra of TGM2 that were obtained by Q-Exactive, coupled with nanoflow capillary high-performance liquid chromatography after trypsin digestion of spot 1 (a) is shown. d Protein binding to GX1 during co-immunoprecipitation was separated by SDS-PAGE and subsequently incubated with TGM2-antibody. Results of Western blotting showed that a specific band was detected in IP-GX1 (approximately 100 kDa, indicated by red arrow 4) lane and that no band was detected at the same position in IP-URP lane