Abstract
The volume of molecular observations on human diseases in public databases is continuously increasing at accelerating rates. A bottleneck is their computational integration into a coherent description, from which researchers may derive new well-founded hypotheses. Also, the need to integrate data from different technologies (genetics, coding and regulatory RNA, proteomics) emerged in order to identify biomarkers for early diagnosis and prognosis of complex diseases and therefore facilitating the development of novel treatment approaches. We propose here a workflow for the integrative transcriptomic description of the molecular pathology in Parkinsons’s Disease (PD), including suggestions of compounds normalizing disease-induced transcriptional changes as a paradigmatic example. We integrated gene expression profiles, miRNA signatures, and publicly available regulatory databases to specify a partial model of the molecular pathophysiology of PD. Six genetic driver elements (2 genes and 4 miRNAs) and several functional network modules that are associated with PD were identified. Functional modules were assessed for their statistical significance, cellular functional homogeneity, literature evidence, and normalizing small molecules. In summary, our workflow for the joint regulatory analysis of coding and non-coding RNA, has the potential to yield clinically as well as biologically relevant information, as demonstrated here on PD data.
Introduction
Parkinson’s disease (PD) is a progressive neurodegenerative disorder that affects several regions of the brain, particularly the substania nigra (SN) dopamine neurons controlling balance and movements1,2. It is considered to be the second most frequent neurodegenerative disorder after Alzheimer’s disease with a prevalence of approximately 180 per 100,000 inhabitants leading to ≈1,260,000 affected individuals in the European population3,4. Since both the incidence and prevalence of PD is increasing with age as for all neurodegenerative diseases3, the frequency of PD is continuously increasing in ageing societies, and thus PD and other neurodegenerative diseases become more and more important for their health care systems. The current treatment strategies only include symptomatic therapies using dopamine replacement by levodopa or dopamine agonist administration, antiglutamatergic or anticholinergic strategies or deep brain stimulation, but no causative therapeutic approaches have been identified. To identify biomarkers for early diagnosis and prognosis and to develop innovative causative treatments, detailed knowledge of the genetic causes and major determinants and regulators driving the underlying neurodegenerative process is an obligate prerequisite. However, due to the complex and heterogenic nature of the disease5–7, many of the cellular pathways and molecular regulators underlying PD pathology have not been unraveled so far.
Among the molecular regulators, transcription factors (TFs) and microRNAs (miRNAs) are key players for regulating gene expression8. Together they play crucial roles in regulating cellular processes9, and their malfunction can cause genetic disorders as well as complex diseases such as PD2,10,11. More specifically, TFs and miRNAs frequently form Feed Forward Loops (FFLs) and other network motifs to regulate cellular transcription in a connective manner10,12,13. Therefore, utilizing the combined regulatory information on TFs/genes and miRNAs as well as their target genes can shed light on key driver genes/TFs and miRNAs in human diseases and, in turn, suggest novel therapeutic strategies14.
Recent advances in molecular genetics revealed several factors implicated in the etiology of PD, such as inherited genetic predispositions and environmental factors that lead to acquired alterations15,16. For instance, it was demonstrated that SN neurons of PD patients show aberrant gene expression patterns that affect critical signaling pathways2,17,18. Moreover, miRNA expression analysis of human SN neurons showed distinct profiles that are dysregulated in PD patients2,19,20 and hence suggested critical functional roles of miRNAs in SN neurons as well as in PD pathogenesis as proposed in21,22.
Despite these important findings based on separate analyses of gene and miRNA expression, there is a lack of studies of the associated miRNA-mRNA interactome as well as of their co-regulation mechanisms, and of their collaborative functional roles in the underlying pathogenesis of PD. For instance, Chandrasekaran and co-workers 2013 analyzed several microarray gene expression datasets and biomolecular networks to identify novel genes and miRNAs of relevance to PD23. Also Dong et al. 2016 predicted ten potential therapeutic targets by studying the overlap between the differentially expressed genes and the target genes of the differentially expressed miRNAs in PD samples24. In the context of neurodegenerative diseases in general, we recently established a consensus-based strategy to integrate miRNA and gene expression data in multiple sclerosis to unravel the unknown cellular roles of miRNAs in this neuroimmune disease25.
As of today, the community has not yet agreed on best practices for such a joint analysis. A particular difficulty lies in the identification of gene-regulatory players and processes, especially since the control units of miRNA expression levels and the miRNAs they control were often found on different chromosomes26. This is a distinct difference with respect to coding RNA, which more frequently features cis-acting control27. An effective joint analysis will hence require the integration of a series of tools with tailored parameters for the interpretation of each dataset to reveal the regulatory mechanisms of disease pathogenesis.
To this end, we propose a comprehensive workflow for integrating miRNA expression profiles, gene expression profiles, and publicly available regulatory databases, in order to gain a deeper understanding of the co-regulation mechanisms and the collaborative functional role of miRNAs and genes in driving disease processes (see Fig. 1). The present study applied the workflow to PD as a paradigmatic example and by doing this yielded 19 dysregulated miRNAs and 116 dysregulated genes that may contribute to the molecular alterations behind PD. Out of these dysregulated genes and miRNAs, 24 genes and 3 miRNAs were found to be well known biomarkers in PD. Furthermore, we constructed a combinatorial gene regulatory network and highlighted six central hub nodes (two dysregulated genes and four dysregulated miRNAs) that could act as PD drivers; further experimental research is warranted to confirm these findings. Next, we identified recurring network motifs (functional modules) of dysregulated genes/TFs and miRNAs and co-targeted genes and we validated the cooperative functional role of their elements in the regulatory activities during PD pathogenesis in terms of statistical significance and biological relevance. Finally, we characterized small compounds and neuroprotective drugs that may normalize the regulatory activity of these functional modules.
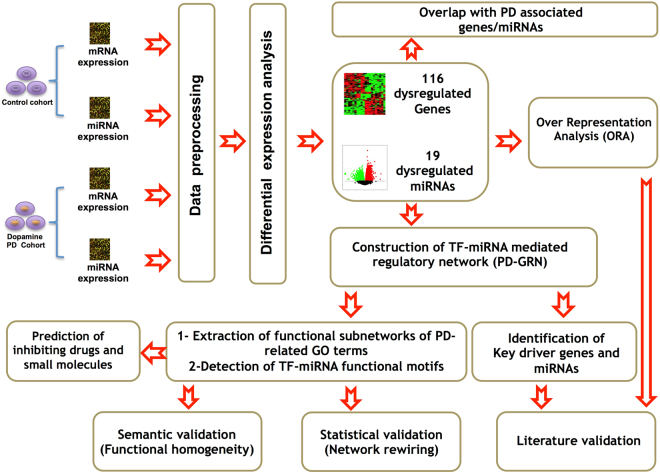
Figure 1.
A schematic diagram for the integrative transcriptomic workflow. The sketch describes data processing and integration of two different transcriptomic datasets to detect major determinants and functional modules controlling PD.
Results and Discussion
Workflow description
We developed and applied an integrative bioinformatics workflow to conduct an integrative analysis with the aim of identifying the major genetic drivers and co-operative functional network modules that describe the molecular pathology of the disease and hence suggest disease-modulating drug compounds (Fig. 1). The workflow starts out with the separate analyses of expression data of mRNAs and miRNAs, including tests for differential expression. The differential expression data are then subject to combinatorial network analysis for the identification of hub nodes (i.e. candidate mRNAs/miRNAs that may drive the disease) as well as important functional network modules. The descriptive transcriptomics part concludes with a listing of these candidate genes and functional modules in the context of the current insights of the studied disease. We extended the workflow for an automated selection of drugs from LINCS28 and the Connectivity Map (CMap)29 with predicted normalizing effects on the dysregulated functional modules which may drive the disease. Formerly, we successfully confirmed the usefulness of multiple components of the workflow by applying it to a large number of breast cancer samples derived by different expression technologies to unravel the complex regulatory architecture of breast invasive carcinoma13. Apart from disease pathogenesis but in the context of cellular processes, we utilized the workflow in part to explain how imprinted genes contribute to cellular differentiation processes and to developmental stages of hematopoiesis30. Here we demonstrate the efficacy of the workflow in describing the molecular pathology of Parkinson’s disease, a disease with a limited number of biological samples.
Differential analysis and enrichment analysis
We processed gene and miRNA expression datasets for 10 early-onset PD samples and 9 control samples of healthy tissues from previous PD studies (see Methods). These studies adopted separate gene or miRNA expression analyses and provided evidence for correlations of genes or miRNAs with signaling pathways relevant to PD pathogenesis2,19. Here we were concerned with the integrative analysis of both miRNA and gene expression datasets. PCA analysis of the normalized gene expression data clustered the analyzed samples into the corresponding groups (PD/Control) with only one exception per group (see Fig. 2). These two samples may be mislabeled. Nevertheless, they had only negligible impact on the results, as demonstrated when excluding them.
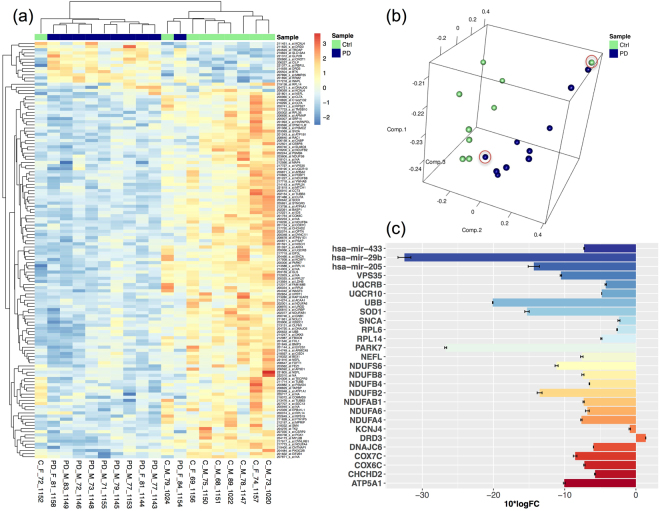
Figure 2.
Differential analysis of gene expression and sample clustering. (a) The heatmap of the expression patterns of the 116 identified dysregulated genes between the PD and the control cohorts. Blue spots represent down-regulation whereas red-yellow spots denote up-regulation patterns. The dendrograms on the upper and left sides show the hierarchical clustering tree of genes or samples. (b) The PCA clustering for the normalized gene expression samples. The two highlighted PD and control samples are incorrectly clustered to the corresponding cohort, however they had almost no impact on the analysis results when we excluded them. (c) The Log fold change (LFC) of the 24 dysregulated genes and the 3 dysregulated miRNAs, which are known to be highly associated with PD progression and pathways. Each colour refers to a different gene or miRNA. The heatmap and PCA clustering for miRNA samples are shown in Figure S1.
The differential expression analysis of the gene and miRNA expression data resulted in 116 dysregulated genes and 19 dysregulated miRNAs, respectively. Figure 2a shows the heatmaps for the relative expressions values of the dysregulated genes between the early-onset PD samples and the control samples. Interestingly, among these dysregulated genes and miRNAs, the genes: PARK7, SNCA, VPS35, CHCHD2, UBB, NEFL, SOD1, KCNJ4, RPL14, RPL6, NDUFA4, NDUFB4, NDUFB8, NDUFA6, NDUFAB1, NDUFB2, COX7C, COX6C, NDUFS6, UQCR10, ATP5A1, UQCRB, DNAJC6, DRD3, and the miRNAs: hsa-mir-433, hsa-mir-205, hsa-mir-29b were found to be strongly associated with PD pathways as annotated in DisGeNET31, OMIM32, HMDD33, or KEGG34 databases, and the list of genes is congruent with the list of genes described in35. All these genes and miRNAs were markedly downregulated in the PD samples except for the DRD3 gene, which was upregulated, see Fig. 2c. The heatmap visualization of the dysregulated miRNAs and the PCA clustering for the miRNA samples are depicted in Figure S1.
The postulated functional roles of the dysregulated genes and miRNAs were backed up by inspecting the associated GO terms and KEGG pathways via overrepresentation analysis (ORA). We identified the most significant functional categories and processes that were enriched in the dysregulated miRNA and gene sets, demonstrating relevance to the etiology of PD, see Tables 1 and S1, respectively. Notably, neurological GO terms and pathways were among the top over-represented entries. For instance, the dysregulated genes were enriched with the functional terms GO:0006119 “oxidative phosphorylation” (p = 1.08E−5), GO:0001963 “synaptic transmission, dopaminergic” (p = 0.001), GO:0048699 “Generation of neurons” (p = 0.002), and the signaling pathways of each of Parkinson’s disease (p = 2.09E−6), Huntington’s disease (p = 1.62E−8), and Alzheimer disease (p = 4.22E−8). Similarly, the dysregulated miRNAs were found to be enriched with cell death function (p = 0.005), neurodegenerative diseases (p = 0.019), and Parkinson’s disease (p = 0.04), indicating the cellular functional roles of the identified dysregulated genes and miRNAs in PD SA neurons.
Table 1.
Enrichment of functional terms, diseases, and tissue specificity within the dysregulated miRNAs.
| Category | Term | Count | adj p.value | miRNAs |
|---|---|---|---|---|
| Function | Epithelial-mesenchymal transition | 4 | 0.0399 | hsa-let-7b,hsa-mir-370,hsa-mir-382,hsa-mir-205 |
| Function | Cell death | 6 | 0.0059 | hsa-let-7b,hsa-mir-130b,hsa-mir-129-5p,hsa-mir-205,hsa-mir-885-5p,hsa-mir-212 |
| Disease | Heart Failure | 9 | 0.0027 | hsa-let-7b,hsa-mir-520d-5p,hsa-mir-129-5p,hsa-mir-139-3p,hsa-mir-382,hsa-mir-205,hsa-mir-130b,hsa-mir-212,hsa-mir-29b |
| Disease | Leukemia Myeloid Acute | 5 | 0.0064 | hsa-let-7b,hsa-mir-323-3p,hsa-mir-382,hsa-mir-29b,hsa-mir-370 |
| Disease | Neurodegenerative Diseases | 2 | 0.0199 | hsa-let-7b,hsa-mir-139-3p |
| Disease | Parkinson’s Disease | 2 | 0.0481 | hsa-mir-433,hsa-mir-29b |
| Disease | Stomach Neoplasms | 5 | 0.0297 | hsa-mir-129-5p,hsa-mir-139-3p,hsa-mir-212,hsa-mir-433,hsa-mir-130b |
| TissueSpecific | Adrenal | 2 | 0.0145 | hsa-mir-370,hsa-mir-485-5p |
| TissueSpecific | Brain | 4 | 0.0007 | hsa-mir-323-3p,hsa-mir-383,hsa-mir-433,hsa-mir-129-5p |
Construction of TF-miRNA mediated regulatory network
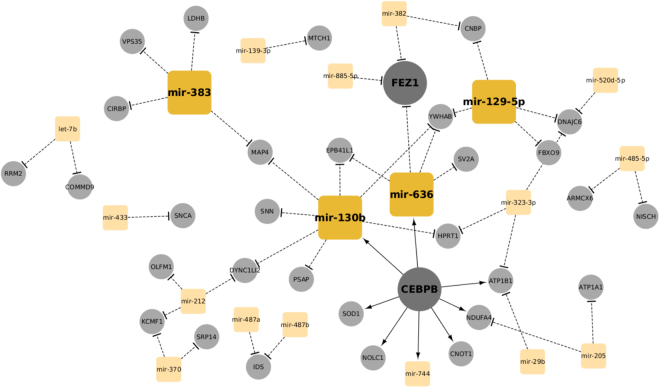
Next we constructed a GRN network (PD-GRN) that combines transcriptional and post-transcriptional regulatory interactions between the dysregulated genes and miRNAs (see Methods). The GRN network contains three different types of interactions: dysregulated gene/TF regulating a dysregulated gene (TF → mRNA), dysregulated miRNA regulating a dysregulated gene (miRNA → mRNA), and dysregulated gene/TF regulating a dysregulated miRNA (TF → miRNA), describing how miRNAs are an essential part of a complex regulation system in PD pathology. In order to characterize the central hub nodes that contribute a large amount of the overall regulation, we computed the node degree centrality parameters and ranked the nodes according to their degrees. We identified 4 central hub miRNAs (hsa-mir-130b, hsa-mir-636, hsa-mir-383, hsa-mir-129-5p) and 2 hub genes (CEBPB, and FEZ1) (Fig. 3). These hub nodes correspond to highly central nodes in the PD-GRN and play a critical role in maintaining the interactions between these genes and their neighborhood genes. Hence, they are candidates for master regulatory genes or for essential genetic drivers, and potential targets for new drugs and treatment of PD. Remarkably, the gene CEBPB was previously identified as a critical central hub node in a protein-protein interaction network related to neuron function activity in PD patients36. CEBPB is also early regulated by CREB37, which is one of the key transcription factors mediating adaptive responses of neurons to bioenergetics challenges38. Many studies have also reported the aberrant expression patterns of the FEZ1 gene and its role in the regulation of the neuronal microenvironment during the progression of PD39,40. Also, expression disruption of the identified key miRNAs hsa-mir-130b, and hsa-mir-636 has been connected to pathogenesis in neuropsychiatric and other neurodegenerative disorders41–44.
Figure 3.
The PD gene regulatory network (PD-GRN) constructed from the dysregulated genes and miRNAs. Large nodes represent key driver genes and miRNAs. Square orange nodes denote the miRNAs, whereas the circular grey nodes represent genes. The network was visualized using the Cytoscape tool.
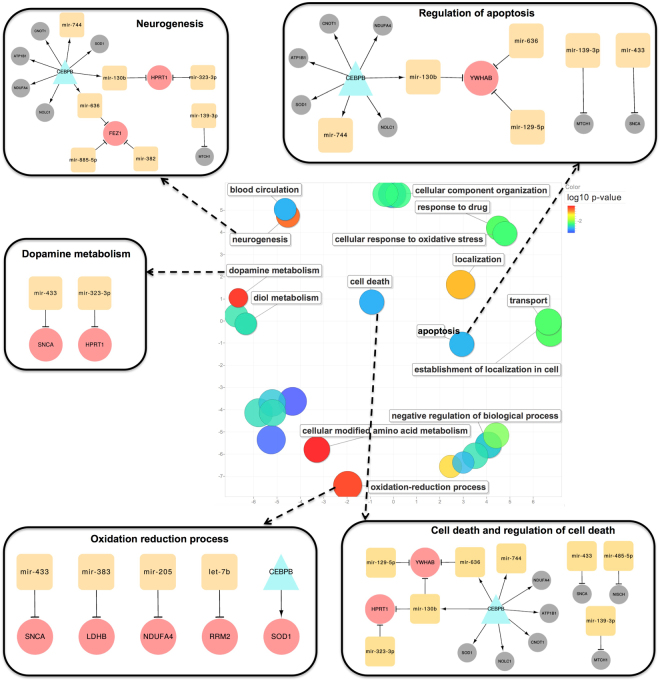
Subsequently, an enrichment analysis on all nodes of the PD-GRN network using DAVID was performed. We discovered a total of 64 enriched GO terms (adjusted p-value < 0.05) that are mostly neurology-related (see Table S2). Guided by the PD GO annotation project (a comprehensive resource providing GO annotation to PD-relevant human gene products)45, we found five important PD-related GO terms that are closely related to PD, in our enrichment list (Fig. 4). We further inspected the association of miRNAs in the PD-GRN to these five GO terms in order to associate specific miRNAs to possible dysregulated pathways in PD. Therefore, for each of the five PD-related functional terms, we created a subnetwork module using the genes associated with these enriched terms as well as their neighboring nodes (Fig. 4). For example, the cell death subnetwork is created from the genes belonging to the enriched GO terms “cell death” and “regulation of cell death”, and their direct miRNA neighbors. There are eight miRNAs in this subnetwork and they regulate both the genes belonging to the enriched GO terms as well as other neighboring nodes. This functional subnetwork therefore provides us with dysregulated pathways concerning cell death in PD. From Fig. 4, we observe that SOD1, CEBPB, and SCNA are annotated with four out of the five GO terms, suggesting that these genes are apparently implicated in various dysregulated pathways in PD. These findings are also supported by recent studies showing the potential roles of SCNA46,47, SOD148, and CEBPB2,36 in the neuronal loss in the PD brain and in PD development. Additionally, the miRNAs mir-130, mir-636, and mir-744 are involved in subnetworks created from enriched GO terms corresponding to abnormal adult neurogenesis, apoptosis, and cell death. This hints at the role of these miRNAs in PD pathogenesis by modulating cell death as well as inhibiting the creation of new functional neuron cells.
Figure 4.
Enrichment analysis of the PD-GRN genes and visualization of five network modules corresponding to PD-related GO terms. Five GO terms are often affected in PD cases. The total list of the enriched GO terms is found in Table S2. The central scatter plot shows the visualization of the top enriched generic GO terms of the PD-GRN in a two dimensional space based on the GO semantic similarities. GO term node colours indicate the p-values for the enrichment of the GO terms. These generic GO terms represent implicitly their subterms, which are not visualized in the plot. The scatter plot was generated using the web tool REVIGO79. All five network modules include both miRNAs and genes. The main TF CEBPB is highlighted by a cyan triangle while the miRNAs are represented by orange squares. The genes co-targeted by TFs and/or miRNAs are depicted in larger pink circles. The regulated genes, regulated by a TF or by a miRNA, are coloured in grey. The network modules were visualized using the Cytoscape tool.
Identification of TF-miRNA co-regulatory motifs and statistical validation
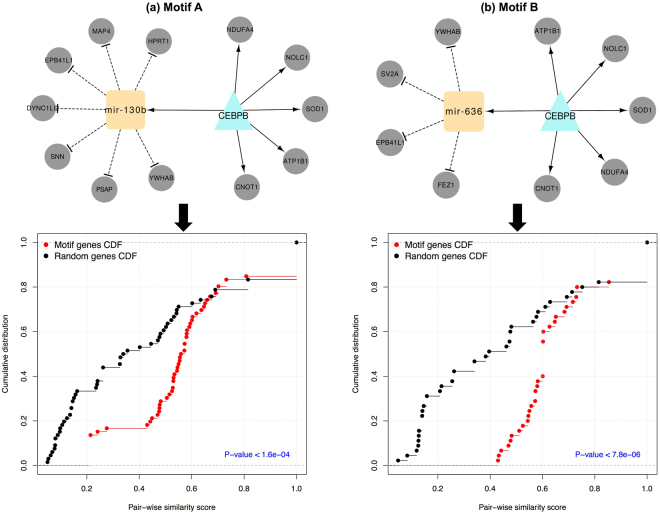
Transcriptional gene regulatory networks often contain functional recurring patterns known as network co-regulatory motifs49 that control multiple features of normal cell function and that may trigger genetic disorders50,51. Here, our workflow checks for the presence of 3-node motifs involving the dysregulated genes and the dysregulated miRNAs in the PD-GRN network (see Methods). We also considered the motif types that were previously described in52. We unveiled a total of 11 cascaded-miRNA-mediated motifs comprising distinct combinations of a TF, a miRNA, a target gene, and co-regulated genes (Table S3). The statistical significance of the motifs was tested by comparing their counts in the network under investigation to their counts in randomized variants of these networks preserving the same node degrees (p-value = 0.03). Interestingly, the 11 motifs included mainly the TF CEBPB (see above) as a main regulator and varied between the two miRNAs hsa-mir-130b and hsa-mir-636 as well as various target genes (Table S3). Therefore, we reduced the 11 motifs to two major motifs (Motif A and Motif B) as shown in Fig. 5.
Figure 5.
The reduced visualization of the detected motifs in the PD-GRN network. Motifs A and B and associated functional homogeneity plots depicting the cumulative distribution of GO functional semantic scores of gene pairs of co-regulated genes in the examined motif (red) versus randomly selected genes (black). The p-value was calculated using the Kolmogorov-Smirnov test. The network motifs were visualized using the Cytoscape tool.
Specifically, motif A, which involves the TF CEBPB, the miRNA hsa-mir-130 and various target genes, is an example of how co-regulatory network motifs may help to better understand the pathogenicity of PD. Intriguingly, the CEBPB gene is involved in PD-related regulatory interactions via binding to the proximal promoter of the ATXN gene (which is the spinocerebellar Ataxia protein associated with the phenotypic variability of neurodegenerative diseases53) and thereby up-regulating its expression in neuroblastoma cells54. The ATXN transcript did not show dysregulation. However, Lee and colleagues 2008 demonstrated that hsa-mir-130 regulates ATXN protein levels in human cells and its inhibition enhances the cytotoxicity caused by the ATXN protein55,56. This sheds light on the potential collaborative role between the hub gene CEBPB and the hub miRNA hsa-mir-130 and their co-regulated genes/miRNAs in regulating ATXN. This strongly confirms our findings and implies that the identified motifs represent functional network modules that might be therapeutic targets for PD.
Semantic assessment: functional homogeneity within the motif nodes
Next, the biological evidence for the two co-regulatory motifs A and B is evaluated in more depth to better assess their cooperative functional roles during the etiology of early-onset PD. We measured the functional homogeneity within each motif by calculating the functional similarity scores between all gene pairs and comparing the resulting distribution to the similarity score distribution of randomly selected gene pairs from the network (p-values < 1.6E-4, Kolmogorov-Smirnov test), see Fig. 5. Intriguingly, the motif genes have significantly more cellular functional homogeneity than randomly selected ones. Therefore, these motifs could highlight new insights into TF-miRNA network modules in PD development, by demonstrating the cooperative functional role between the TFs/genes and their potential miRNA partners, yielding a better understanding of the dysregulation mechanisms involving PD pathways.
Finally, we merged the two main motifs A and B to obtain a consolidated functional network module with their co-regulated genes that are enriched in the PD-GRN, see Figure S2.
Identification of normalizing small molecules and drugs
Our workflow concludes by proposing small molecule interventions that possibly normalize the expression signatures of the merged functional module (Figure S2) using the library of integrated network-based cellular signatures (LINCS) L1000 data set28,57, by identifying (based on the complete extrapolated set of genes, not the landmark genes only) small molecules causing transcriptional changes inversely correlating with our expression signatures. We used an optimized PD signature (the merged functional network module) instead of using all differentially expressed genes or a whole genome expression profile as a signature for the LINCS query with the aim of obtaining superior performance for compound prioritization and assessing the biological relevance of the identified functional modules. Interestingly, LINCS identified known neuroprotective agents (e.g., Staurosporine and Brivanib) and signaling pathway inhibitors (e.g., GDC-0068 and Torin-2), which work on the central nervous system to prevent neuron degradation58 and to delay progression of PD59 and Alzheimer60 in animal models, see Table S4. This supports the hypothesis that the identified functional network modules are relevant for PD, and may open up new avenues for therapy.
Subsequent to the LINCS methodology, observed changes in gene expression were compared against the drug effects described by the CMap29 to identify the drugs that may reverse the whole genome expression profile of PD cases. For each of the 3203 gene expression profiles in the CMap, we generated Rank-rank hypergeometric overlap (RRHO) maps61, showing the extent and pattern of its global similarity to our inverted PD gene expression profile, see Figure S3. We were interested especially in CMap profiles that feature upregulation of genes that are downregulated in the PD profile (i.e. at the top of the list in the inverted PD profile). In a RRHO map such behavior would materialize as a region of high intensity (reflecting low overlap p-values; red color) in the lower left corner of the map, with lower intensities elsewhere. Thus, we computed for each of the 3203 RRHO maps the degree of similarity (distance) to an artificially constructed reference map showing that intensity distribution and ranked the list of RRHO maps accordingly. Figure S3 depicts a panel of 16 maps from the top of that list, i.e. drugs from the CMap whose upregulated genes show considerable overlap with genes downregulated in PD. Among those are hesperetin and valproic acid which were found to have neuroprotective effects by attenuating behavioral abnormality in hemiparkinsonian rats62, and by reversing the alpha-synuclein alterations in a rotenone rat model of PD63, respectively.
Comparison to similar workflows/approaches
Several approaches have been implemented to facilitate the integrative analysis of gene and miRNA expression profiles. For instance, the MMIA web tool integrates miRNA and mRNA expression data using straightforward inverse correlation between the mis-regulated genes and miRNAs as well as gene set enrichment analysis to characterize diseases and pathways related to miRNAs64. The MAGIA65 pipeline provides a higher diversity by allowing the combination of miRNA-target gene predictions for either matched or un-matched miRNA–gene expression profiles using different relatedness measures to end up with a regulatory network for associated phenotypes. Also miRTrail66 performs ORA and Gene Set Enrichment analyses of interactions of genes and miRNAs based on expression profiles. However, it explores only miRNA → gene interactions. DisTMGneT67 was developed for obtaining a cancer-specific network based on expression profiles of dysregulated genes and miRNAs. However, it is limited to a predefined set of miRNAs and genes as well as cancer. Compared to the aforementioned approaches, our workflow has distinctive features of downstream analysis such as identification of driver genes/miRNAs, detecting effective functional network modules, small molecule predictions, and a variety of validation methods. Table S5 summarizes the comparison between our approach and the aforementioned approaches/tools.
Conclusions
We propose a workflow for the joint analysis of coding and non-coding RNA taken from the same samples. It has the potential to yield clinically as well as biologically relevant information, as demonstrated here on PD data, with a very limited number of samples and previously on breast cancer data with a large number of samples. Furthermore, it offers descriptions of distinct molecular processes that are associated with the disease, provides consistent enrichments of Gene Ontology terms and of disease pathways, and identifies cooperative functional modules. For each functional module a network of interacting coding and non-coding transcripts is offered. The resulting data are also subjected to a comparison with drug effect databases to give researchers the opportunity to identify hypotheses for curative effectors. We consider that this workflow nicely links clinical studies (from which the expression data may be derived) back to preclinical research. The presented workflow represents an effective model for the integrative analysis of multiple molecular datasets from different experimental assays. Further work to increase the modularity of this workflow is warranted to ease the exchange of tools for the generation of networks and to foster reusability.
Methods
Material collections and experimental procedures
Genetic material collection, DNA preparation, and microarray experimental procedures for gene and miRNA expression in midbrain dopamine neurons were previously described in the two reference publications for the data we utilized2,19. These authors obtained frozen tissue blocks from the Harvard Brain Tissue Resource Center, containing SN from 9 control subjects and 10 idiopathic PD samples, which were cut using a Microm HM 560 CryoStar cryostat (8 μm), mounted on LEICA Frame Slides with a PET-membrane (1.4 μm), dehydrated, subjected for RNA extraction, followed by hybridization to the HU-133A arrays (Affymetrix, Santa Clara, CA). From these 19 samples, 8 control and 8 PD samples were also used for miRNA profiling using the Human MicroRNA TaqMan qRT-PCR Arrays A v1 or 2.0 (Life Technologies, Foster City, CA, USA).
Pre-processing and differential analysis
The raw expression datasets were normalized using quantile normalization and log2 transformed. The differential expression analysis was performed as previously described68 using three methods: (1) ANOVA test69, (2) moderated t-test70, (3) area under the curve of the receiver operator characteristics (AUC ROC)70. P-values were adjusted using the Benjamini-Hochberg71 procedure to limit the false discovery rate to 5%. Genes/miRNAs that were classified as differentially expressed by at least two of the three methods were included in the list of differentially expressed genes/miRNAs. Raw and preprocessed data are provided as a supplementary file.
Construction of the PD-GRN network and the co-regulatory motifs
The regulatory interactions between the differentially expressed (DE) genes and the DE miRNAs, which were identified in this work, were collected from the TFmiR regulatory databases13. We considered all interactions that are supported by experimental and/or by predicted evidence in this analysis. Driver genes/miRNAs (hub nodes) were identified by determining the highly central nodes in the constructed PD-GRN network. For this, we calculated the degree centrality measure for the PD-GRN network using the R package igraph72 and we selected the top 10% (of all genes and miRNAs) only to foster the consistency in their association to GO biological processes73. Functional network modules (three-node TF-miRNA co-regulatory network motifs consisting of a miRNA, a TF, and a joint target gene) were characterized using the computational procedure described in the TFmiR web server publication13. Cytoscape V3.374 was used to visualize the PD-GRN network and the identified network motifs.
Assessment of driver genes and functional network modules
The following 4-step procedures were used to assess and validate our results:
Significance of the identified network motifs (Statistical validation)
To assess the significance of each motif type, we used the same procedure that we previously developed in13. Briefly, we compared the motif occurrences in the real network to their occurrences in randomized ensembles of these networks with preserved node degrees (number of permutations = 100). Only motifs having p-value < 0.05 were considered for further analysis.
Functional homogeneity within the motif genes (Semantic validation)
We used the GoSemSim R package75 to estimate semantic similarity scores according to the Gene Ontology (GO) annotations. Statistical significance was performed by comparing the similarity scores of the motif genes to the similarity scores of randomly selected genes (with the same number of the motif genes). The permutation procedure was repeated 100 times. Then, the Kolmogorov-Smirnov test was adopted to check whether the similarity scores of motif gene pairs were statistically higher than the scores of randomly selected pairs.
Over-representation analysis for genes and miRNAs
Over-representation analysis (ORA) of the miRNA sets was performed using TAM web service76 which identifies the functional classes and disease terms that are enriched in a miRNA set. For the gene sets, the DAVID77 tool was utilized to identify the GO terms which are annotated to at least two genes and are statistically overrepresented in the DE genes as previously shown in reference78. For both genes and miRNA enrichment analysis, Fisher’s Exact test was performed followed by the Benjamini–Hochberg71 adjustment for controlling the false discovery rate (FDR), with a cutoff value of 0.05.
Identifying inhibiting small molecules and drugs
-
For each functional module expression profile
We downloaded the complete version of the library of integrated network-based cellular signatures (LINCS) L1000 data set28,57 on level 3 from the GEO repository (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE70138) and computed the log2 fold change (LFC) of all L1000 genes. The PD-related LFC of the genes involved in the functional network modules was subtracted from the same genes in the processed LINCS data and then summed across all genes in the module to end up with a representative score for each drug. Low-scoring drugs feature a maximum desired expression change on the entire gene set of the functional network module. The drug scores are sorted ascendingly and the first ten drugs were considered to provide biologists/clinicians with a limited but potentially worthwhile set of drug predictions.
-
For the whole genome PD expression profile
The genes in the PD gene expression profile were ranked based on the LFC, from the most downregulated gene to the most upregulated one (inverted profile). We also ranked the genes in each of the 3203 drug gene expression profiles from the ConnectivityMap (CMap)29 based on the log2 fold change from the most upregulated gene to the most downregulated one. For every pair of a PD profile and a CMap profile we computed a rank-rank hypergeometric overlap (RRHO) map61. A desirable match between the PD profile and a given CMap profile would result in a RRHO map with a characteristic intensity distribution. We constructed an artificial map depicting such an intensity distribution as a positive reference and compared the list of RRHO maps with it. All maps were normalized, serialized into a vector and column-wise assembled into a matrix. We performed Principal Component Analysis (PCA) using the rows of that matrix as features and determined the number of principal components (PCs) needed to account for 90% of the total variance. Using those PCs, we calculated a weighted Euclidean distance for each map to the reference map, with the weight reflecting the amount of explained variance for each PC. Finally, we ranked the RRHO maps by their distance to the reference map.
Workflow reusability
This integrative workflow can be classified into two main software modules:
the differential analysis, concerning the identification of the differentially expressed genes/miRNAs. This module was developed as an R script and can be downloaded from the Bitbucket repository (https://hamed20@bitbucket.org/hamed20/differential-analysis.git). The outputs are tab-delimited files of differentially expressed genes/miRNAs, which can be used as input to the next module.
the combinatorial analysis, including the other workflow components. This analysis is accessible using our public webservice TFmiR: http://service.bioinformatik.uni-saarland.de/tfmir. The user guides are included in the corresponding software modules.
Availability Data and Materials
Raw and pre-processed data are available as supplementary.
Electronic supplementary material
Acknowledgements
We thank Kai Sonntag (Harvard Medical School) for providing the raw data for the miRNA analysis. M.H. and G.F. thank Azza Hamed and Yasmeen Qawashmeh for the technical assistance. This work was supported by the BMBF, VIP - Validierung des Innovationspotentials wissenschaftlicher Forschung (03V0396) and the EU (“Aging with elegans”, Grant agreement No 633589).
Author Contributions
M.H. conceptualized the study, developed the workflow, and originally drafted the manuscript. Y.G. contributed to data visualization and expression analysis. S.F., and M.E. participated in predicting the normalizing drugs. S.S. and S.M. revised and advised on the analysis and the manuscript draft. A.S. helped with conceptualizing the study and critically revised the manuscript. G.F. directed the study and helped to draft the manuscript. All authors read and approved the final draft.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-25754-5.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Braak H, Del Tredici K. Invited Article: Nervous system pathology in sporadic Parkinson disease. Neurology. 2008;70:1916–1925. doi: 10.1212/01.wnl.0000312279.49272.9f. [DOI] [PubMed] [Google Scholar]
- 2.Briggs CE, et al. Midbrain dopamine neurons in Parkinson’s disease exhibit a dysregulated miRNA and target-gene network. Brain Research. 2015;1618:111–121. doi: 10.1016/j.brainres.2015.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.von Campenhausen S, et al. Prevalence and incidence of Parkinson’s disease in Europe. European Neuropsychopharmacology. 2005;15:473–490. doi: 10.1016/j.euroneuro.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 4.Riedel O, et al. Die Epidemiologie des idiopathischen Parkinson-Syndroms und assoziierter Demenz und Depression in Dresden. Fortschritte der Neurologie· Psychiatrie. 2013;81:81–87. doi: 10.1055/s-0032-1330278. [DOI] [PubMed] [Google Scholar]
- 5.Schrag A, Quinn NP, Ben-Shlomo Y. Heterogeneity of Parkinson’s disease. Journal of Neurology, Neurosurgery & Psychiatry. 2006;77:275–276. [PMC free article] [PubMed] [Google Scholar]
- 6.Sharma M, et al. Large-scale replication and heterogeneity in Parkinson disease genetic loci. Neurology. 2012;79:659–667. doi: 10.1212/WNL.0b013e318264e353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klein C, Schlossmacher MG. The genetics of Parkinson disease: implications for neurological care. Nature Clinical Practice Neurology. 2006;2:136–146. doi: 10.1038/ncpneuro0126. [DOI] [PubMed] [Google Scholar]
- 8.Kuijjer, M. L., Glass, K. & Quackenbush, J. Gene regulation by transcription factors and microRNAs in ovarian cancer. Cancer Research75, 10.1158/1538-7445.compsysbio-b1-20 (2015).
- 9.Zhou, Y., Ferguson, J., Chang, J. T. & Kluger, Y. Inter-and intra-combinatorial regulation by transcription factors and microRNAs. Bmc Genomics8, 10.1186/1471-2164-8-396 (2007). [DOI] [PMC free article] [PubMed]
- 10.Poos, K. et al. How MicroRNA and Transcription Factor Co-regulatory Networks Affect Osteosarcoma Cell Proliferation. Plos Computational Biology9, 10.1371/journal.pcbi.1003210 (2013). [DOI] [PMC free article] [PubMed]
- 11.Elstner M, et al. Expression analysis of dopaminergic neurons in Parkinson’s disease and aging links transcriptional dysregulation of energy metabolism to cell death. Acta Neuropathol. 2011;122:75–86. doi: 10.1007/s00401-011-0828-9. [DOI] [PubMed] [Google Scholar]
- 12.Yan, Z. et al. Integrative analysis of gene and miRNA expression profiles with transcription factor-miRNA feed-forward loops identifies regulators in human cancers. Nucleic Acids Research40, 10.1093/nar/gks395 (2012). [DOI] [PMC free article] [PubMed]
- 13.Hamed M, Spaniol C, Nazarieh M, Helms V. TFmiR: a web server for constructing and analyzing disease-specific transcription factor and miRNA co-regulatory networks. Nucleic Acids Res. 2015;43:W283–288. doi: 10.1093/nar/gkv418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamed, M., Spaniol, C., Zapp, A. & Helms, V. Integrative network-based approach identifies key genetic elements in breast invasive carcinoma. Bmc Genomics16, 10.1186/1471-2164-16-s5-s2 (2015). [DOI] [PMC free article] [PubMed]
- 15.Schapira AH, Jenner P. Etiology and Pathogenesis of Parkinson’s Disease. Movement Disorders. 2011;26:1049–1055. doi: 10.1002/mds.23732. [DOI] [PubMed] [Google Scholar]
- 16.Wirdefeldt K, Adami H-O, Cole P, Trichopoulos D, Mandel J. Epidemiology and etiology of Parkinson’s disease: a review of the evidence. European Journal of Epidemiology. 2011;26:S1–S58. doi: 10.1007/s10654-011-9581-6. [DOI] [PubMed] [Google Scholar]
- 17.Cantuti-Castelvetri I, et al. Effects of gender on nigral gene expression and parkinson disease. Neurobiology of Disease. 2007;26:606–614. doi: 10.1016/j.nbd.2007.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simunovic, F., Yi, M., Wang, Y., Stephens, R. & Sonntag, K. C. Evidence for Gender-Specific Transcriptional Profiles of Nigral Dopamine Neurons in Parkinson Disease. Plos One5, 10.1371/journal.pone.0008856 (2010). [DOI] [PMC free article] [PubMed]
- 19.Kim W, et al. miR-126 contributes to Parkinson’s disease by dysregulating the insulin-like growth factor/phosphoinositide 3-kinase signaling. Neurobiology of Aging. 2014;35:1712–1721. doi: 10.1016/j.neurobiolaging.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim W, et al. MiR-126 Regulates Growth Factor Activities and Vulnerability to Toxic Insult in Neurons. Molecular Neurobiology. 2016;53:95–108. doi: 10.1007/s12035-014-8989-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wong G, Nass R. miRNAs and their putative roles in the development and progression of Parkinson’s disease. Frontiers in genetics. 2012;3:315–315. doi: 10.3389/fgene.2012.00315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heman-Ackah, S. M., Hallegger, M., Rao, M. S. & Wood, M. J. A. RISC in PD: the impact of microRNAs in Parkinson’s disease cellular and molecular pathogenesis. Frontiers in Molecular Neuroscience6, 10.3389/fnmol.2013.00040 (2013). [DOI] [PMC free article] [PubMed]
- 23.Chandrasekaran S, Bonchev D. A Network View on Parkinson’s Disease. Computational and Structural Biotechnology Journal. 2013;7:e201304004. doi: 10.5936/csbj.201304004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dong N, Zhang X, Liu Q. Identification of therapeutic targets for Parkinson’s disease via bioinformatics analysis. Mol Med Rep. 2017;15:731–735. doi: 10.3892/mmr.2016.6044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Freiesleben, S., Hecker, M., Zettl, U. K., Fuellen, G. & Taher, L. Analysis of microRNA and Gene Expression Profiles in Multiple Sclerosis: Integrating Interaction Data to Uncover Regulatory Mechanisms. Scientific Reports6 (2016). [DOI] [PMC free article] [PubMed]
- 26.Gupta Y, et al. Dissecting genetics of cutaneous miRNA in a mouse model of an autoimmune blistering disease. BMC Genomics. 2016;17:112. doi: 10.1186/s12864-016-2455-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jansen RC, Nap JP. Genetical genomics: the added value from segregation. Trends Genet. 2001;17:388–391. doi: 10.1016/S0168-9525(01)02310-1. [DOI] [PubMed] [Google Scholar]
- 28.Duan QN, et al. LINCS Canvas Browser: interactive web app to query, browse and interrogate LINCS L1000 gene expression signatures. Nucleic Acids Research. 2014;42:W449–W460. doi: 10.1093/nar/gku476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lamb J. Innovation - The Connectivity Map: a new tool for biomedical research. Nature Reviews Cancer. 2007;7:54–60. doi: 10.1038/nrc2044. [DOI] [PubMed] [Google Scholar]
- 30.Hamed M, et al. Linking Hematopoietic Differentiation to Co-Expressed Sets of Pluripotency-Associated and Imprinted Genes and to Regulatory microRNA-Transcription Factor Motifs. PLoS One. 2017;12:e0166852. doi: 10.1371/journal.pone.0166852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pinero, J. et al. DisGeNET: a discovery platform for the dynamical exploration of human diseases and their genes. Database-the Journal of Biological Databases and Curation, 10.1093/database/bav028 (2015). [DOI] [PMC free article] [PubMed]
- 32.Hamosh A, Scott AF, Amberger JS, Bocchini CA, McKusick VA. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Research. 2005;33:D514–D517. doi: 10.1093/nar/gki033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu, M. et al. An Analysis of Human MicroRNA and Disease Associations. Plos One3, 10.1371/journal.pone.0003420 (2008). [DOI] [PMC free article] [PubMed]
- 34.Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Research. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simunovic F, et al. Gene expression profiling of substantia nigra dopamine neurons: further insights into Parkinson’s disease pathology. Brain. 2009;132:1795–1809. doi: 10.1093/brain/awn323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao L, Gao H, Zhou H, Xu Y. Gene expression profiling analysis of the putamen for the investigation of compensatory mechanisms in Parkinson’s disease. BMC Neurol. 2013;13:181. doi: 10.1186/1471-2377-13-181. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Papadia S, et al. Synaptic NMDA receptor activity boosts intrinsic antioxidant defenses. Nat Neurosci. 2008;11:476–487. doi: 10.1038/nn2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Camandola, S. & Mattson, M. P. Brain metabolism in health, aging, and neurodegeneration. EMBO J, 10.15252/embj.201695810 (2017). [DOI] [PMC free article] [PubMed]
- 39.Sun YY, et al. Fasciculation and elongation protein zeta-1 (FEZ1) expression in reactive astrocytes in a rat model of Parkinson’s disease. Neuropathol Appl Neurobiol. 2014;40:164–176. doi: 10.1111/nan.12077. [DOI] [PubMed] [Google Scholar]
- 40.Desplats P, et al. Combined exposure to Maneb and Paraquat alters transcriptional regulation of neurogenesis-related genes in mice models of Parkinson’s disease. Mol Neurodegener. 2012;7:49. doi: 10.1186/1750-1326-7-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maffioletti E, Tardito D, Gennarelli M, Bocchio-Chiavetto L. Micro spies from the brain to the periphery: new clues from studies on microRNAs in neuropsychiatric disorders. Frontiers in Cellular Neuroscience. 2014;8:16. doi: 10.3389/fncel.2014.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Provost P. MicroRNAs as a molecular basis for mental retardation, Alzheimer’s and prion diseases. Brain Research. 2010;1338:58–66. doi: 10.1016/j.brainres.2010.03.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lau P, et al. Alteration of the microRNA network during the progression of Alzheimer’s disease. Embo Molecular Medicine. 2013;5:1613–1634. doi: 10.1002/emmm.201201974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu QY, et al. Identification of microRNAs involved in Alzheimer’s progression using a rabbit model of the disease. Am J Neurodegener Dis. 2014;3:33–44. [PMC free article] [PubMed] [Google Scholar]
- 45.Foulger RE, et al. Using the Gene Ontology to Annotate Key Players in Parkinson’s Disease. Neuroinformatics. 2016;14:297–304. doi: 10.1007/s12021-015-9293-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Siddiqui IJ, Pervaiz N, Abbasi AA. The Parkinson Disease gene SNCA: Evolutionary and structural insights with pathological implication. Sci Rep. 2016;6:24475. doi: 10.1038/srep24475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oczkowska A, Kozubski W, Lianeri M, Dorszewska J. Mutations in PRKN and SNCA Genes Important for the Progress of Parkinson’s Disease. Curr Genomics. 2013;14:502–517. doi: 10.2174/1389202914666131210205839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trist BG, et al. Amyotrophic lateral sclerosis-like superoxide dismutase 1 proteinopathy is associated with neuronal loss in Parkinson’s disease brain. Acta Neuropathol. 2017;134:113–127. doi: 10.1007/s00401-017-1726-6. [DOI] [PubMed] [Google Scholar]
- 49.Shen-Orr SS, Milo R, Mangan S, Alon U. Network motifs in the transcriptional regulation network of Escherichia coli. Nature genetics. 2002;31:64–68. doi: 10.1038/ng881. [DOI] [PubMed] [Google Scholar]
- 50.He, L. et al. A microRNA component of the p53 tumour suppressor network. Nature447, 1130–1134 (2007). [DOI] [PMC free article] [PubMed]
- 51.Li X, Cassidy JJ, Reinke CA, Fischboeck S, Carthew RW. A microRNA imparts robustness against environmental fluctuation during development. Cell. 2009;137:273–282. doi: 10.1016/j.cell.2009.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li K, et al. Functional analysis of microRNA and transcription factor synergistic regulatory network based on identifying regulatory motifs in non-small cell lung cancer. BMC Syst Biol. 2013;7:122. doi: 10.1186/1752-0509-7-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang Y, et al. Genetic and clinical analysis in a Parkinson’s disease family caused by expansion of SCA2. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2015;32:776–779. doi: 10.3760/cma.j.issn.1003-9406.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 54.Chen IC, et al. ATXN8-62 G/A promoter polymorphism and risk of Taiwanese Parkinson’s disease. European Journal of Neurology. 2012;19:1462–1469. doi: 10.1111/j.1468-1331.2012.03749.x. [DOI] [PubMed] [Google Scholar]
- 55.Lee Y, et al. miR-19, miR-101 and miR-130 co-regulate ATXN1 levels to potentially modulate SCA1 pathogenesis. Nat Neurosci. 2008;11:1137–1139. doi: 10.1038/nn.2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Meza-Sosa KF, Valle-Garcia D, Pedraza-Alva G, Perez-Martinez L. Role of microRNAs in central nervous system development and pathology. Journal of Neuroscience Research. 2012;90:1–12. doi: 10.1002/jnr.22701. [DOI] [PubMed] [Google Scholar]
- 57.Wang ZC, Clark NR, Ma’ayan A. Drug-induced adverse events prediction with the LINCS L1000 data. Bioinformatics. 2016;32:2338–2345. doi: 10.1093/bioinformatics/btw168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gitto, S. B. Recent insights into the pathophysiology of mTOR pathway dysregulation (2015).
- 59.Wright JW, Kawas LH, Harding JW. A Role for the Brain RAS in Alzheimer’s and Parkinson’s Diseases. Frontiers in Endocrinology. 2013;4:158. doi: 10.3389/fendo.2013.00158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Patel NS, et al. Alzheimer’s beta-amyloid peptide blocks vascular endothelial growth factor mediated signaling via direct interaction with VEGFR-2. J Neurochem. 2010;112:66–76. doi: 10.1111/j.1471-4159.2009.06426.x. [DOI] [PubMed] [Google Scholar]
- 61.Plaisier SB, Taschereau R, Wong JA, Graeber TG. Rank-rank hypergeometric overlap: identification of statistically significant overlap between gene-expression signatures. Nucleic Acids Res. 2010;38:e169. doi: 10.1093/nar/gkq636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Baluchnejadmojarad, T. & Roghani, M. The Flavonoid Hesperetin Alleviates Behavioral Abnormality in 6-Hydroxydopamine Rat Model of Hemi-Parkinsonism. Basic and Clinical Neuroscience2 (2010).
- 63.Monti B, et al. Valproic acid is neuroprotective in the rotenone rat model of Parkinson’s disease: involvement of alpha-synuclein. Neurotox Res. 2010;17:130–141. doi: 10.1007/s12640-009-9090-5. [DOI] [PubMed] [Google Scholar]
- 64.Nam S, et al. MicroRNA and mRNA integrated analysis (MMIA): a web tool for examining biological functions of microRNA expression. Nucleic Acids Research. 2009;37:W356. doi: 10.1093/nar/gkp294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sales G, et al. MAGIA, a web-based tool for miRNA and Genes Integrated Analysis. Nucleic Acids Research. 2010;38:W352. doi: 10.1093/nar/gkq423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Laczny C, et al. miRTrail-a comprehensive webserver for analyzing gene and miRNA patterns to enhance the understanding of regulatory mechanisms in diseases. BMC bioinformatics. 2012;13:36. doi: 10.1186/1471-2105-13-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sengupta D, Bandyopadhyay S. Topological patterns in microRNA–gene regulatory network: studies in colorectal and breast cancer. Molecular bioSystems. 2013;9:1360–1371. doi: 10.1039/c3mb25518b. [DOI] [PubMed] [Google Scholar]
- 68.Hamed M, Spaniol C, Zapp A, Helms V. Integrative network-based approach identifies key genetic elements in breast invasive carcinoma. BMC Genomics. 2015;16(Suppl 5):S2. doi: 10.1186/1471-2164-16-S5-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tsai CA, Chen YJ, Chen JJ. Testing for differentially expressed genes with microarray data. Nucleic Acids Res. 2003;31:e52. doi: 10.1093/nar/gng052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hahne, F., Huber, W., Gentleman, R. & Falcon, S. Bioconductor case studies (Springer, 2010).
- 71.Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat Med. 1990;9:811–818. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
- 72.Csardi G, Nepusz T. The igraph software package for complex network research. InterJournal, Complex Systems. 2006;1695:1–9. [Google Scholar]
- 73.Nazarieh M, Wiese A, Will T, Hamed M, Helms V. Identification of key player genes in gene regulatory networks. BMC Syst Biol. 2016;10:88. doi: 10.1186/s12918-016-0329-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Smoot ME, Ono K, Ruscheinski J, Wang P-L, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yu G, et al. GOSemSim: an R package for measuring semantic similarity among GO terms and gene products. Bioinformatics. 2010;26:976–978. doi: 10.1093/bioinformatics/btq064. [DOI] [PubMed] [Google Scholar]
- 76.Lu M, Shi B, Wang J, Cao Q, Cui Q. TAM: a method for enrichment and depletion analysis of a microRNA category in a list of microRNAs. BMC bioinformatics. 2010;11:419. doi: 10.1186/1471-2105-11-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2008;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 78.Hamed M, Ismael S, Paulsen M, Helms V. Cellular Functions of Genetically Imprinted Genes in Human and Mouse as Annotated in the Gene Ontology. PloS one. 2012;7:e50285. doi: 10.1371/journal.pone.0050285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Supek F, Bošnjak M, Škunca N, Šmuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PloS one. 2011;6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.