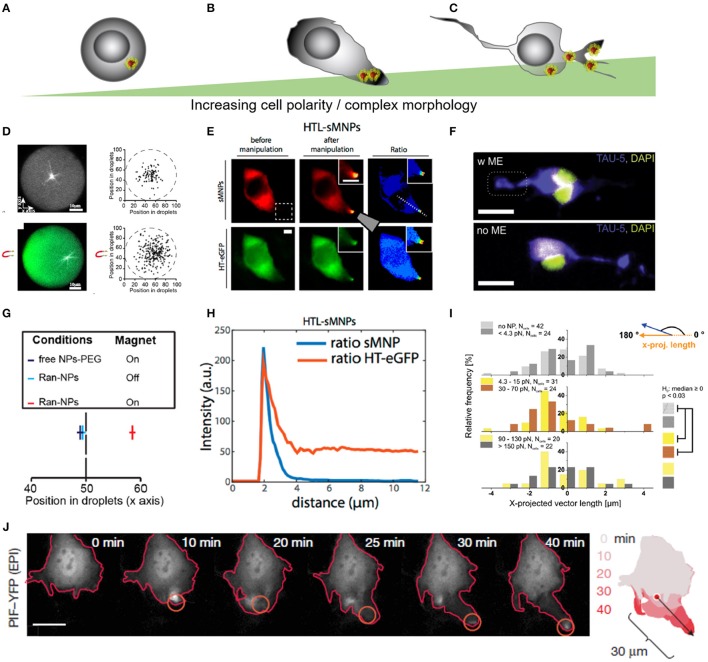
Figure 5.
Force-mediated protein sorting inside cells across different levels of complexity in cell morphology. (A–C) Schematic representation of different levels of cell complexity ranging from almost perfectly round to highly branched structures. (D) Microtubules nucleation in artificial, micro-scaled lipid droplets de-centralize the nucleation zone through the application of magnetic field gradient. Magnetic forces off-center nucleation position of microtubules through repulsion. Forces were estimated in the femtonewton range. (E) Altered protein positioning (HaloTag-eGFP) through nanomagnetic forces operated by magnetic tweezers in HeLa cells. Scale bar = 10 μm. HTL-sMNPs, HaloTag-ligand-silica-based magnetic nanoparticles. (F) Primary cortical neurons with superparamagnetic nanoparticles re-assemble Tau proteins toward the magnetic field gradient when exposed to a permanent magnetic field. Scale bar = 16 μm. (G) Microtubules nucleation position of RanGTP-magnetic nanoparticles (Ran-NPs) without (Off) and with (On) magnetic forces. (D,G) Reproduced with permission from Bonnemay et al. (2013), Copyright © 2013, American Chemical Society. (H) Surface intensity plot shows correlation between nanoparticles (HTL-sMNPs = sMNPs) and protein assembly (HT-eGFP) in transfected HeLa cells dropping away from the magnetic tip. (E,H) Reproduced with permission from Etoc et al. (2015), Copyright © 2015, American Chemical Society. (I) Histogram plot for the nanomagnetic force range were protein assembly was significant different from its native distribution. (F,I) Adapted from Kunze et al. (2015), Copyright © 2015, American Chemical Society. (J) Force-mediated local activation actin cytoskeleton dynamics through dragging Rho-family GTPases proteins. Reproduced with permission from Levskaya et al. (2009), Copyright © 2009, Springer Nature.