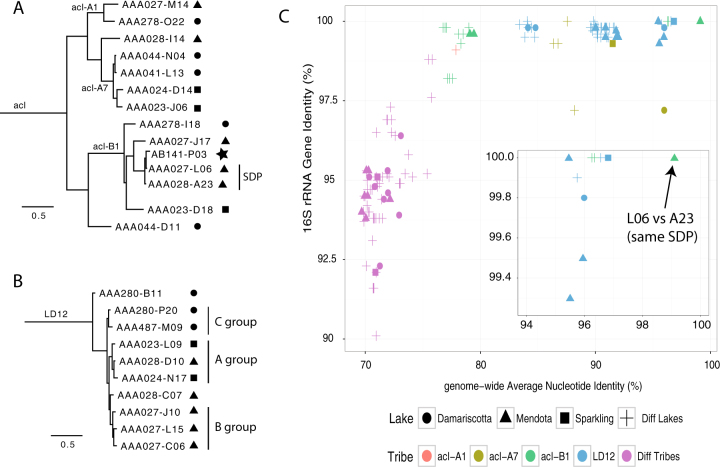
Fig. 1.
a Phylogenetic tree of acI SAGs based on conserved single copy genes selected by PhyloPhlAn. Amino acid sequences from 400 genes were aligned. The tree topology is consistent with 16S rRNA gene-based phylogenies [30]. SAGs L06 and A23 are part of the same sequence-discrete population (SDP) as defined in the text and further based on data shown in Fig. 2. b Phylogenetic tree of LD12 SAGs based on conserved single copy genes selected by PhyloPhlAn, representing 400 genes. The tree topology was consistent with prior work that provided evidence for finer-scale groups within the LD12 tribe [31]. c Genome-wide nucleotide identity (gANI) vs. 16S rRNA gene identity for pairs of SAGs. Alignment fractions for homologous genomic regions and 16S rRNA genes are given in Table S2. Shapes indicate the lake the tribe is from, if same, otherwise different lake is indicated. Colors indicate the tribe a pair is from, if same, otherwise different tribe is indicated. The arrow denotes the L06–A23 pair