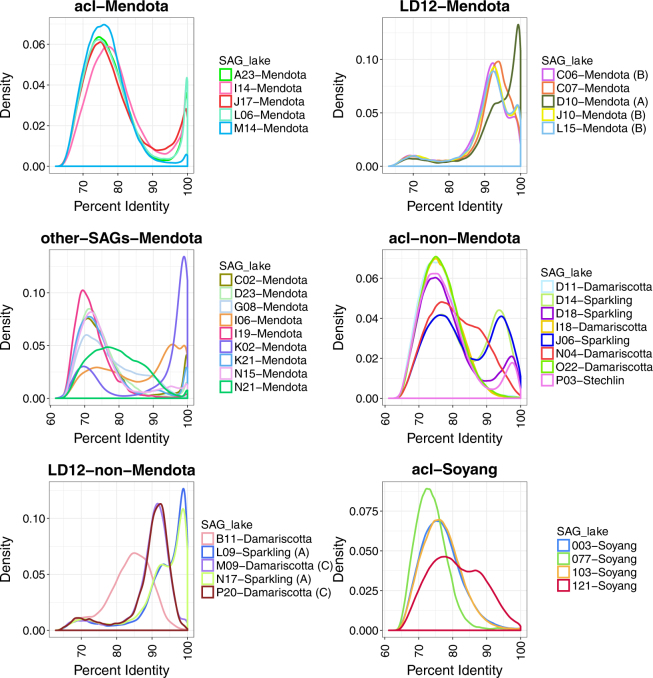
Fig. 3. Mapping metagenomic reads from Lake Mendota to SAGs and four genomes from Lake Soyang.
[27]. The x-axis represents nucleotide identity of the recruited reads. The metagenome sample was collected from Lake Mendota on 29 April 2009. Reads were only counted if they aligned over a minimum of 200 bp. Recruitments were not competitive, meaning that each read could recruit to multiple SAGs. Analogous competitive recruitments that required each read to recruit to only one SAG are presented in Fig. S4. The non-competitive recruitment showed the close relationship of the LD12 populations that is not visible in the competitive recruitment. An expanded multi-panel version of the same data is presented in Fig. S3 for clarity. Each panel represents a different subset of the SAGs: a acI from Mendota, b acI not from Mendota, c LD12 from Mendota (group members demarcated in legend), d LD12 not from Mendota (group members demarcated in legend), e other freshwater groups from Mendota, f genomes from Lake Soyang, Korea. Regarding the other freshwater groups from Mendota, since each of these SAGs represent just one tribe, it is not appropriate to infer any general conclusions for these populations or tribes, but we present them here to show the intriguing diversity of recruitment patterns. We finally underscore the need to more deeply sample individual population members using SAGs, to better capture and describe the range of variation in population heterogeneity