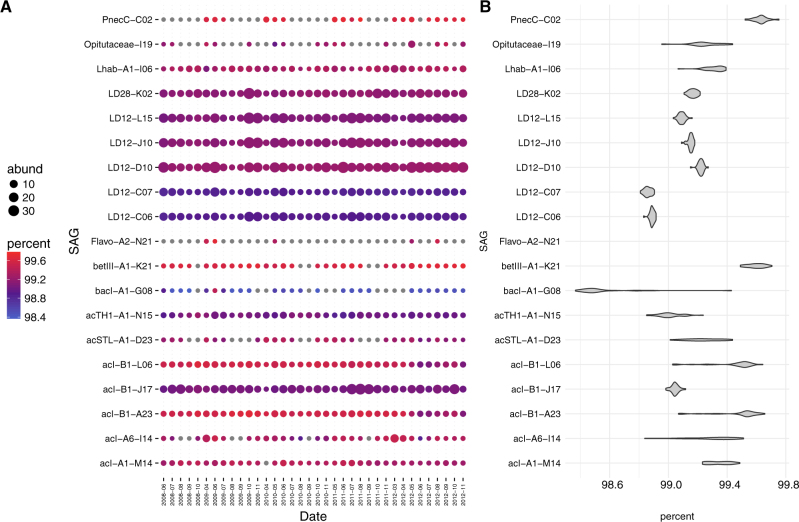
Fig. 5.
a Metagenomic read recruitment using the SAGs from Lake Mendota. SAGs are in rows with bubbles representing all metagenomes from a particular month recruited against SAG. Filtering criteria: ≥97.5% ANI and ≥200 bp alignment length. Color scale indicates the ANI of the recruited metagenome reads. Bubble size represents the average coverage per base in the reference SAG divided by the size of the metagenome, multiplied by the average size of all metagenomes (1.34 Gigabases). Gray bubbles indicate that fewer than 200 reads recruited to the SAG in that month. Note that the resulting values do not represent a true measure of absolute abundance, but allow for quantitative comparison of month-to-month variation in population-level abundance. Recruitments were performed competitively, meaning that each read was counted for only one SAG, unless the read hit two SAGs equally well in which case it was counted for both SAGs. b Variation in ANI for each SAG, across all 30 metagenomes from throughout the five years. Variation was not calculated for a SAG unless at least 10 months recruited more than 200 hits each. The data underlying these plots can be found in Table S6