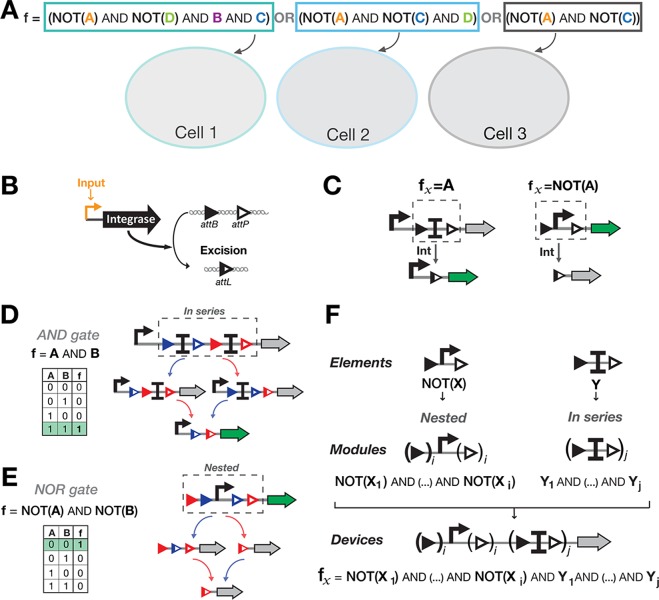
Figure 2.
A hierarchical composition framework for asynchronous Boolean recombinase logic. (A) Distribution of a Boolean function within a multicellular consortium by decomposition into conjunctions of literals (variables or their negations). Here an example is depicted in which a Boolean function is decomposed into three subfunctions and implemented in three separate cellular computing units. (B) attB and attP disposed in parallel orientation. (C) Elements implementing IDENTITY and NOT functions. To obtain an IDENTITY function, a transcriptional terminator is flanked by parallel attachment sites, blocking transcription of the gene of interest. When the signal is present, the terminator is excised and the output gene is expressed. To obtain a NOT function, a promoter is flanked by parallel attachment sites. When the signal is present, the promoter is excised, and the gene is no longer expressed. (D) Functional composition of ID-elements into ID-modules, by placing elements in series to obtain the conjunction of IDENTITY functions. For a 2-input ID-module, the output gene is expressed only when both inputs have been present, both terminators excised (corresponding to an AND gate (A AND B)). (E) Functional composition of NOT-elements into NOT-modules, by nesting elements to obtain conjunction of NOT functions. For a 2-input NOT-module, the output gene is expressed only when none of the inputs has been present (corresponding to a NOR gate: NOT(A) AND NOT(B)). (F) Hierarchical composition framework for Boolean recombinase logic. ID- and NOT-modules are composed in series, following a priority rule in which the NOT-module is placed upstream the ID-module. The device shown here can be scaled to perform all functions based on conjunction of NOT and IDENTITY functions.