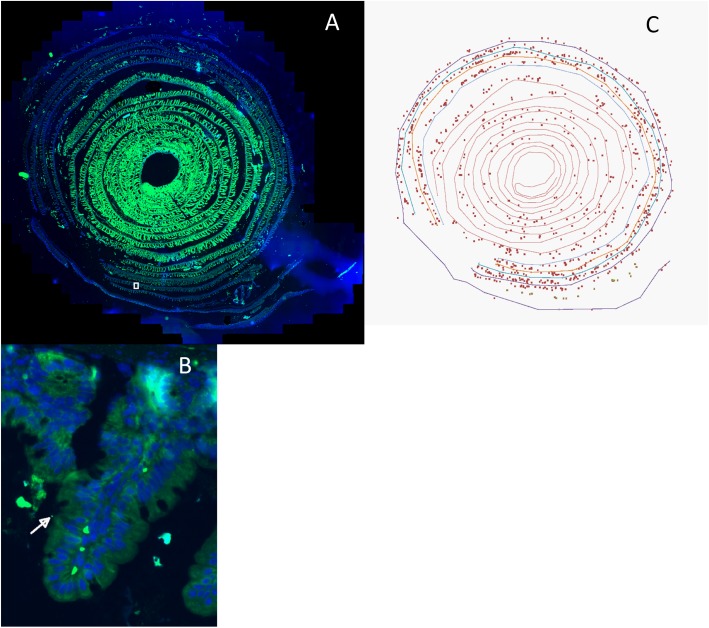
FIGURE 1.
(A) Intestinal “Swiss roll” micrograph of a mouse fed with raw milk cheese contaminated with Enterohemorrhagic Escherichia coli (EHEC) O157:H7 strain EDL933 SR225. The intestinal sample was rolled from duodenum to recto-anal junction so that the anal junction was at the outer edge of the spiral. Intestinal cells were labeled using DAPI (Blue), EHEC cells were labeled using anti-O157 IgM coupled with FITC (Green). White square represents the area zoomed in (B). (B) Micrograph of the zoomed area identified by the white square in (A). Bacteria labeled with FITC and DAPI and attached to intestinal layer where intestinal villi were entirely observable were counted. Arrow shows an example of an enumerated bacterium. (C): The position of each bacterium in picture (A) was identified (xb, yb) and the contour of the basal lumen of the mouse recto-anal junction to duodenum intestinal section was extracted by hand (∼300 points). The position of each bacterium along the intestine was obtained by identifying the point of the contour that was closest to the bacterium and oriented toward the center of the spiral. Micrograph obtained with AxioScan Z1 (Zeiss).