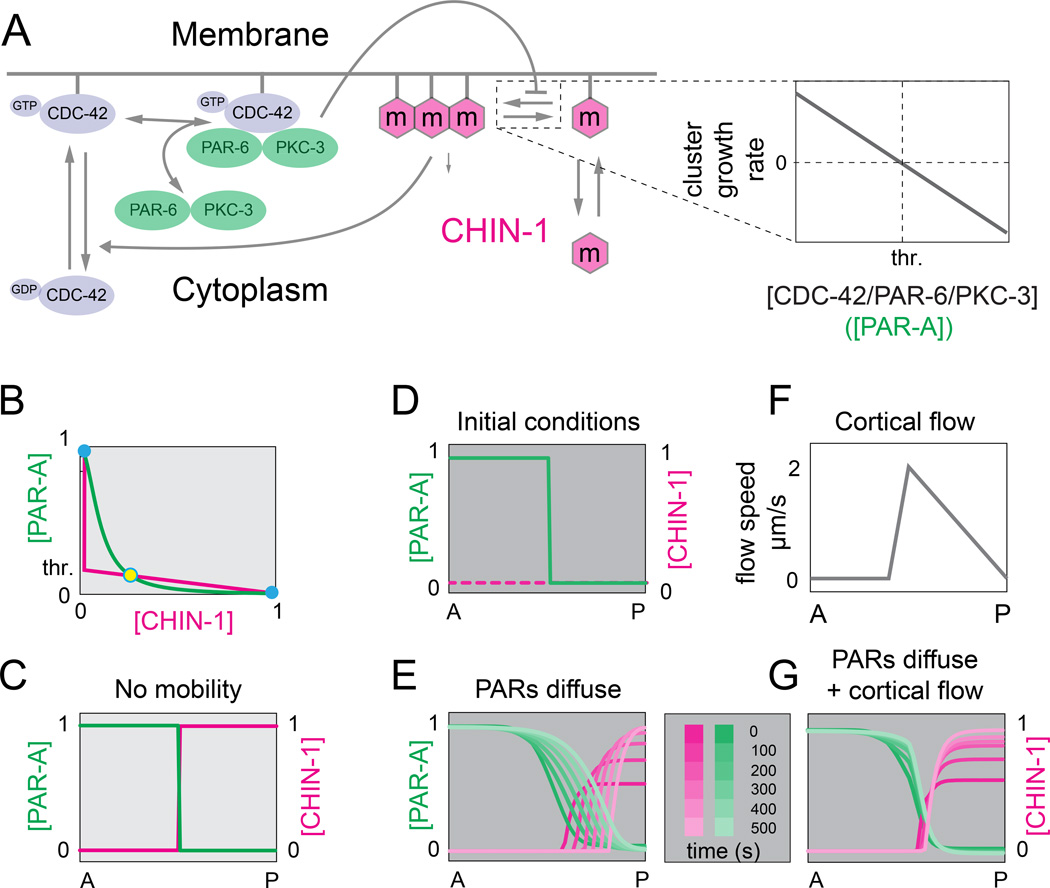
Figure 7. Ultrasensitive dependence of CHIN-1 cluster growth on PAR-6/PKC-3 levels yields bistable dynamics and a stable boundary position.
(A) Schematic view of model assumptions: CDC-42 cycles between inactive and active forms; PAR-6/PKC-3 dimers reversibly bind active CDC-42 from a cytoplasmic pool. CHIN-1 monomers cycle between the cytoplasm and plasma membrane, where they assemble into larger CHIN-1 clusters. CHIN-1 clusters act locally to promote inactivation and dissociation of CDC-42; CDC-42/PAR-6/PKC-3 trimers inhibit local clustering of CHIN-1, such that clusters grow below a threshold level of CDC-42/PAR-6/PKC-3 (expanded view at right; see Supplementary Modeling Procedures for details). (B) Steady state dependence of [CHIN-1] on [PAR-A] (magenta curve), and [PAR-A] on [CHIN-1] (green curve). PAR-A represents the CDC-42/PAR-6/PKC-3 trimer. Steady states occur where the two curves intersect. Solid blue circles indicate stable steady states; open blue circle indicates an unstable steady state. (C) Spatial coexistence of the two stable states in the absence of diffusion. (D) Initial conditions for the simulations shown in E and G. (E) Predicted PAR-A and CHIN-1 distributions vs. time for simulations that incorporate measured values for PAR-6/PKC-3 and CHIN-1 mobility and turnover. (F,G) Predicted PAR-A and CHIN-1 distributions vs. time (G) for simulations that incorporate same parameter values as in (E), and the flow field shown in (F). See Supplemental Modeling Procedures for details. See also Figure S5.