Fig. 3.
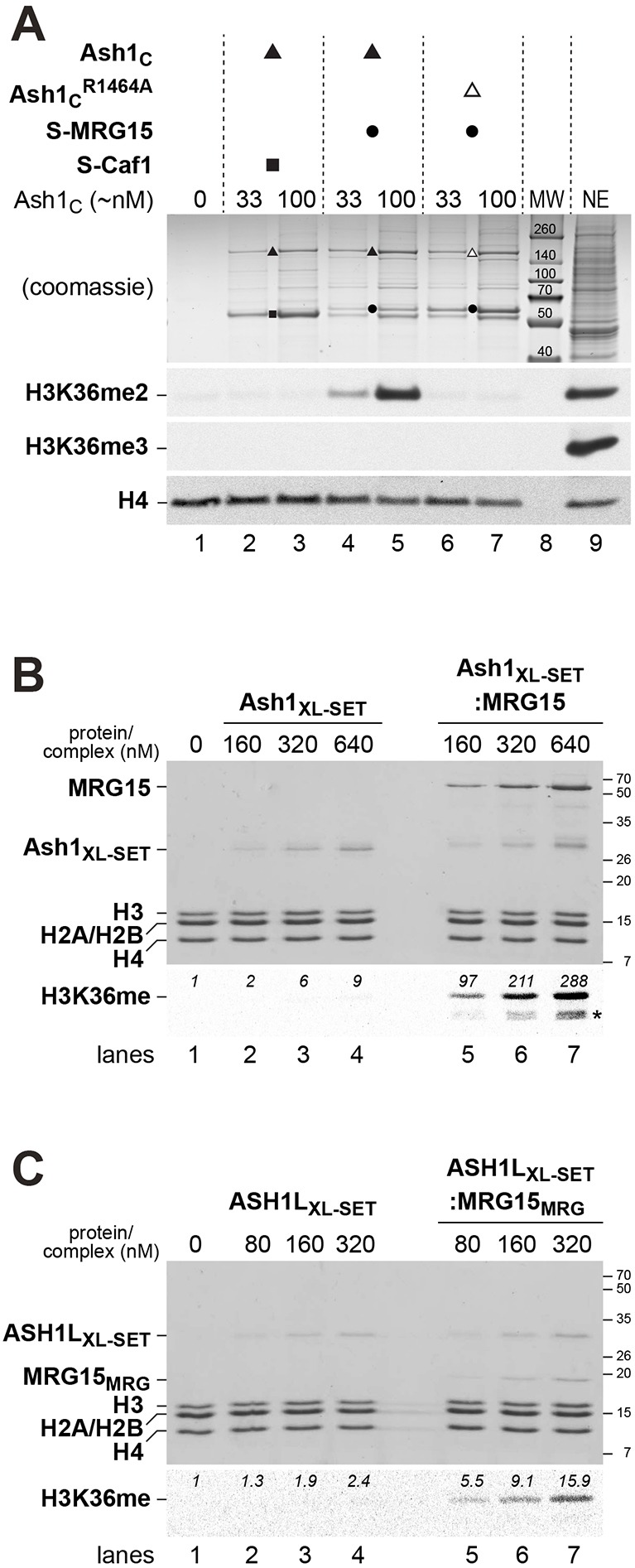
H3K36 di-methylation by Drosophila Ash1 and human ASH1L is stimulated by MRG15. (A) HMTase reactions with recombinant Drosophila Ash1C:Caf1 (lanes 2, 3), Ash1C:MRG15 (lanes 4, 5) or Ash1CR1464A:MRG15 (lanes 6, 7) complexes and reconstituted recombinant mononucleosomes (400 nM in lanes 1-7) were separated on a 10% SDS-polyacrylamide gel; the upper part of the gel was stained with Coomassie Blue to visualize the enzyme complexes, the bottom part was analysed by western blotting with antibodies against H3K36me2 and, as a control, H4. The same reaction was also analysed with antibody against H3K36me3. Enzyme concentrations in the reactions were normalized by estimating Ash1C concentration relative to a Coomassie Blue-stained protein standard. Drosophila embryo nuclear extract (NE) in lane 9 served as control for western blot analysis. Lane 8: molecular weight marker (MW). (B) HMTase reactions with recombinant Drosophila Ash1XL-SET (lanes 2-4) or Ash1XL-SET:MRG15 complex (lanes 5-7) on reconstituted recombinant Xenopus oligonucleosomes (320 nM in lanes 1-7). One part of the reaction was analysed on a 15% SDS-polyacrylamide gel to visualize proteins by Coomassie Blue staining (top), the other part of the reaction was separated on a 15% SDS-polyacrylamide gel, transferred to membrane and analysed by fluorography. PhotoShop software was used to quantify radioactive signal in the H3 band; this signal represents the sum of H3K36me1 and H3K36me2. Asterisk indicates H3 degradation products. (C) HMTase reactions with recombinant human ASH1LXL-SET (lanes 2-4) or ASH1LXL-SET:MRG15MRG complex (lanes 5-7) on reconstituted recombinant Xenopus oligonucleosomes (320 nM in lanes 1-7). Reactions were analysed as in B.
