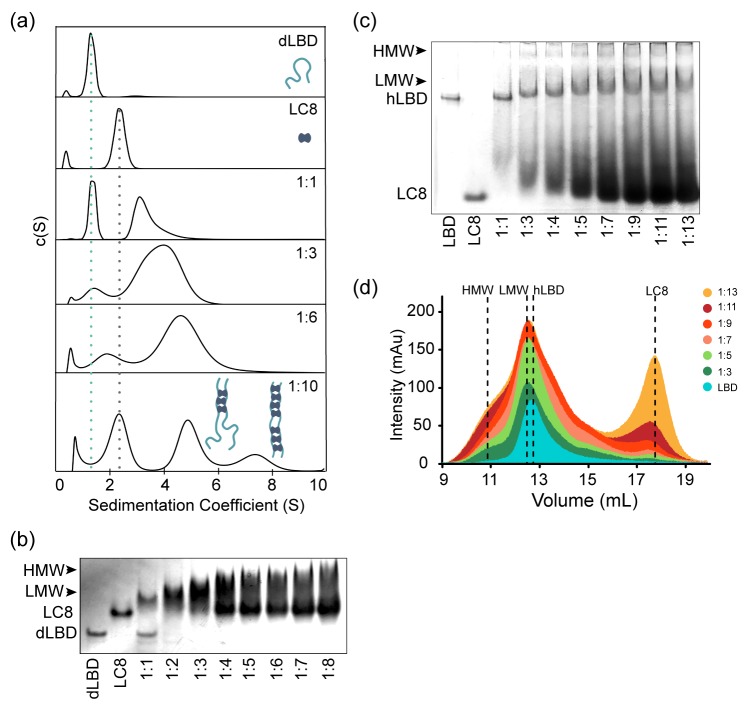
Figure 4. ASCIZ and LC8 form a dynamic complex with low occupancy intermediates.
(a) Representative c(S) distributions obtained by sedimentation velocity are shown for the dLBD, LC8, and increasing molar ratios of the dLBD: LC8; 1:1, 1:3, 1:6, and 1:10. The standardized sedimentation coefficients (s20,w) of unbound LC8 and dLBD determined from sedimentation velocity are 2.42 s (frictional ratio, f/f0 = 1.21, consistent with a globular protein) and 1.34 s (f/f0 = 1.84, consistent with an asymmetric or unfolded protein), respectively. Calculation of the molecular weight from the sedimentation coefficient and frictional ratio gave masses of 23 kDa and 18 kDa respectively, closely matching their theoretical masses of 24 kDa for LC8 dimer and 17 kDa for dLBD monomer. Cartoon depictions of the ASCIZ dLBD (light blue) and LC8 (dark blue) are shown to aid in visualization of the complexes formed at a 1:10 ratio. (b) Native gel titration of dLBD with LC8. An increasing concentration of LC8 was added to a constant amount of dLBD, from a molar ratio of (dLBD:LC8) 1:1 to 1:8. As more LC8 is added, the complex migrates more slowly and excess LC8 appears at 1:4. (c) Native gel titration of hLBD with LC8. As an increasing concentration of LC8 is added to a constant amount of hLBD. Arrows indicate the locations of the low molecular weight (LMW) and high molecular weight (HMW) complex. (d) Titration of the hLBD with LC8, monitored by size exclusion chromatography on a Superdex 200 gel filtration column. The concentration of hLBD is held constant and an increasing amount of LC8 is added, from a molar ratio of 1:3 to 1:13. Peaks corresponding to free hLBD, free LC8, low molecular weight complex (LMW) and high molecular weight (HMW) complex are labeled. Additionally, SAXS data for the LMW dLBD:LC8 complex is shown in Figure 4—figure supplement 1.