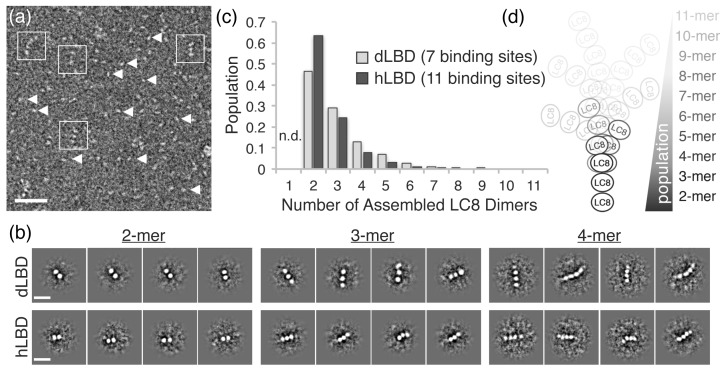
Figure 5. dLBD:LC8 and hLBD:LC8 complexes visualized by negative stain electron microscopy.
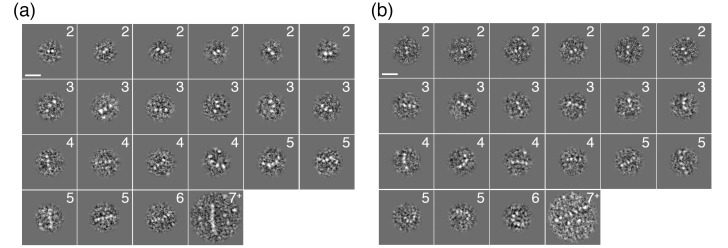
(a) Representative micrograph of negatively stained hLBD:LC8 complexes. Identified oligomeric complexes are boxed. Non-oligomeric LC8 dimers are indicated by arrowheads. Scale bar = 100 nm. dLBD:LC8 micrographs had the same appearance (not shown). (b) Representative 2D projection averages of (top) dLBD:LC8 oligomers and (bottom) hLBD:LC8 oligomers. Only low-occupancy oligomers with 2–4 LC8 dimers (2mer – 4mers) were successfully averaged. Higher-occupancy oligomers were identified in raw micrographs (Figure 5—figure supplement 1), but were not averaged due to low population and/or high degree of conformational flexibility. Scale bar = 20 nm. (c) Histogram showing the normalized population distribution of LC8 occupancy in complexes formed with dLBD (grey) and hLBD (black), identified from raw micrographs. The population of complexes formed with a single LC8 dimer were not determined (n.d.) (d) Illustration representing the distribution of LC8 occupancy and conformational flexibility observed in hLBD:LC8 complexes.