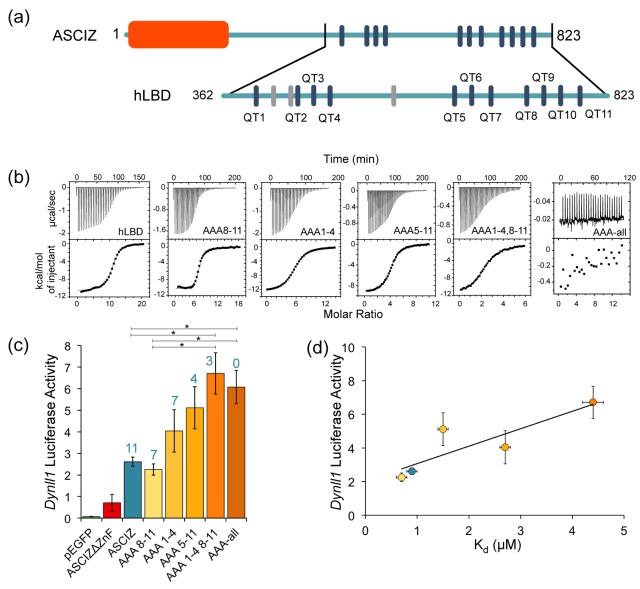
Figure 7. The number of LC8 recognition motifs tunes ASCIZ transcriptional activity.
(a) Domain structure of human ASCIZ, showing 11 LC8 binding motifs as blue bars. Additional non-TQT motifs are shown as gray bars. (b) Shown are representative isothermal titration plots of LC8 with the hLBD and constructs with four (AAA1-4 and AAA8-11), seven (AAA5-11), eight (AAA1-4, 8–11), or fourteen (AAA-all) mutant LC8 binding sites. Data were fit to a single site binding model using Origin software. (c) Firefly luciferase reporter assays of ASCIZ knockout mouse embryo fibroblast cells transiently transfected with WT human ASCIZ (blue), a zinc finger deletion construct (red), and ASCIZ mutant constructs AAA1-4, AAA8-11, AAA5-11, AAA1-4, 8–11, or AAA-all (shades of orange), along with the Dynll1 luciferase and Renilla luciferase vectors. The number of available motifs is indicated above each construct. Error bars are ±S.E. relative to Renilla luciferase as a control. Asterisks (*) indicate p values less than 0.01. Data is the average of 2–4 independent experiments. Although the differences in transcriptional activity between each construct is small, the overall trend indicates an increase in transcriptional activity with a decrease in available binding motifs. A western blot depicting expression of the ASCIZ constructs is shown in Figure 7—figure supplement 1. (d) Binding affinity of each construct for LC8 is plotted against luciferase activity. Data points are colored according to (c). The AAA-all construct is excluded from the graph because it does not bind to LC8.
Figure 7—figure supplement 1. Western blot of ASCIZ constructs.