Figure 3. mRNAseq comparisons between T. gondii and H. hammondi identify unique aspects of the H. hammondi transcriptome.
(A) Venn diagrams indicating the degree of overlap in genes of significantly different transcript abundance between T. gondii and H. hammondi at D4 and D15 post-infection. While overall there are distinct genes that are significantly different at D4 and D15 (Venn diagram, upper left), H. hammondi expresses high numbers of bradyzoite genes even at 4 dPI and this number increases further by D15 (21 additional genes; Venn diagram in upper right). This progression towards an increase in the number of bradyzoite genes over time is not recapitulated among merozoite-specific genes, in that only four additional detectable genes from this gene set were found to be of significantly higher abundance (Padj <0.01; Fold-change ≥4) at 15 DPI. Moreover 18 of the 22 total detectable merozoite genes were of significantly different abundance in H. hammondi, indicating its transcriptional similarity to both bradyzoites and merozoites. (B) Gene Sets found to be significantly (FDR q-value <0.05) enriched in H. hammondi high-abundance transcripts (top) or T. gondii high-abundance transcripts (bottom) at either 4 or 15 days in culture. Gene Set Details can be found in Table 2. *: p<0.05; **: p<0.01; ***: p<0.001. (C) GSEA plots of the in vitro bradyzoite, in vitro tachyzoite, and Cat stage specific one gene sets, showing enrichment profiles between H. hammondi and T. gondii at 15 DPI. NES; Normalized enrichment score. FDR q: False discovery rate q-value. (D) Hierarchically clustered expression data for detectable genes in the CAT STAGE SPECIFIC one gene set in H. hammondi or T. gondii grown for 4 or 15 days in vitro, or for T. gondii grown as tachyzoites or merozoites (in cat enteroepithelial cells).
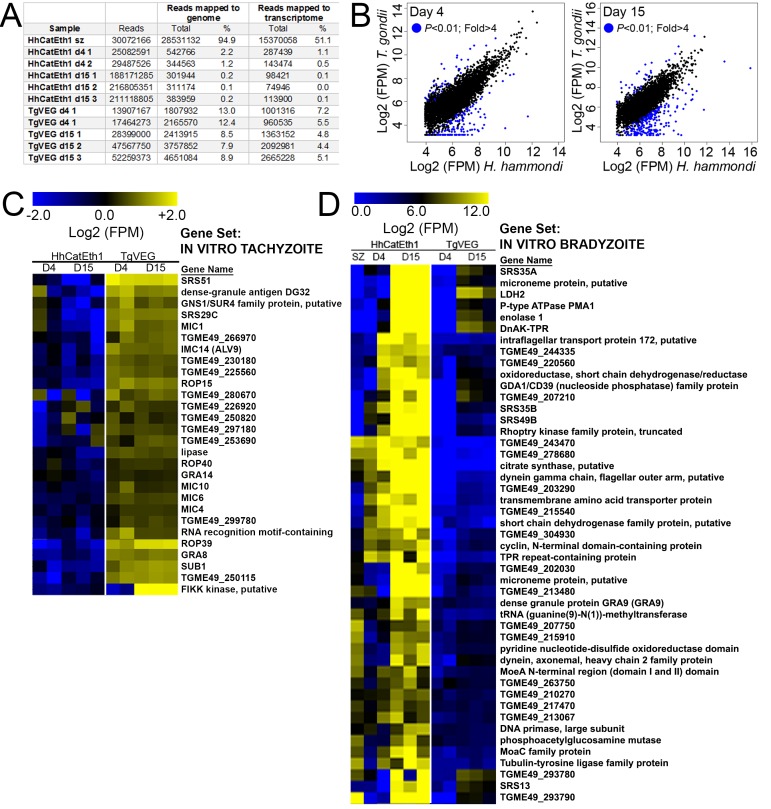
Figure 3—figure supplement 1. Transcriptional profiling and Gene Set Enrichment Analysis of T. gondii and H. hammondi.
Figure 3—figure supplement 2. mRNA-seq comparisons between TgVEG and HhCatAmer sporozoites identify unique H. hammondi transcriptome profiles.