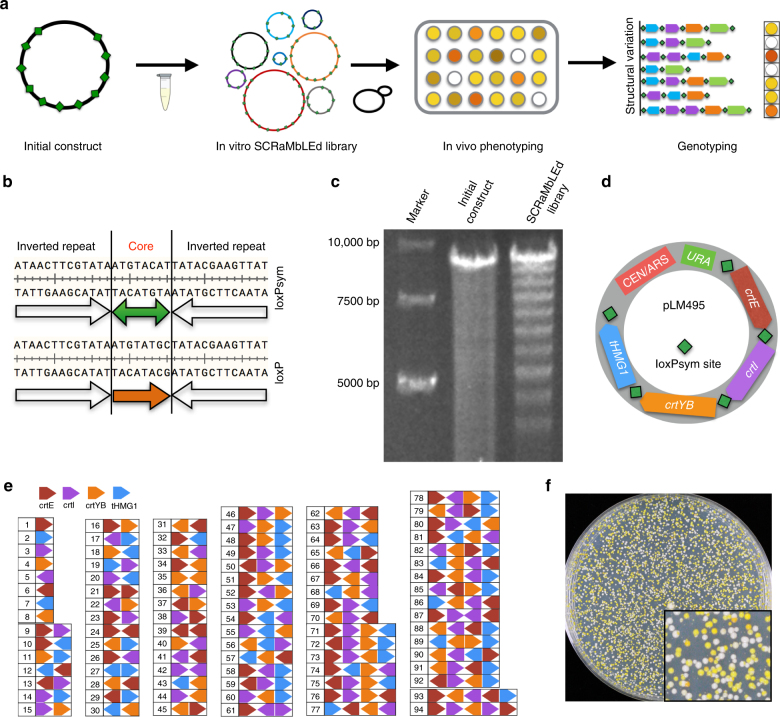
Fig. 1.
Top-down in vitro SCRaMbLE. a Schematic of top-down in vitro SCRaMbLE. Green diamonds represent the 34 bp loxPsym site. b Sequence comparison between loxPsym and loxP sites. c Gel electrophoresis analysis of an in vitro SCRaMbLEd library. The parental construct encoded 10 loxPsym sites with an inter-site distance of 500 bp. Material for linearization with NotI was extracted from a pool of E. coli colonies carrying the SCRaMbLEd DNA. d Map of pLM495. LoxPsym sites flank the β-carotene pathway genes crtE, crtI, crtYB, and tHMG1. Transcription units for these genes are pTIP1-crtE-tACS2, pPGK1-crtI-tASC1, pTDH3-crtYB-tCIT1, pZEO1-tHMG1-tACS2. e A total of 94 unique pathway structures were determined by PacBio sequencing of a SCRaMbLEd pLM495 library. f Yeast colonies transformed with in vitro SCRaMbLEd pLM495. The magnified region shows different colony colors, consistent with production of colored carotenoid intermediates. Synthetic complete medium lacking uracil (SC–Ura) medium was used to select transformants